8Z8M
 
 | |
8Z8V
 
 | |
8JYG
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-(3-phenoxyphenyl)ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based lead optimization to improve potency and selectivity of a novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid series of heparanase-1 inhibitor.
Bioorg.Med.Chem., 93, 2023
|
|
7YI7
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-[4-(trifluoromethyl)phenyl]ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
7YJC
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
6KZX
 
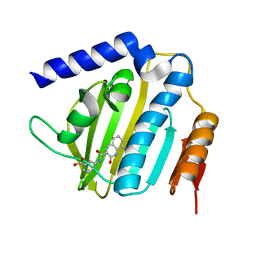 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6L01
 
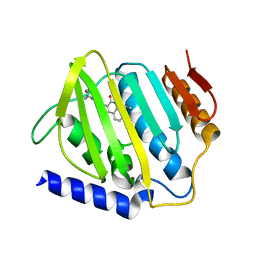 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 2-[3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]phenyl]ethanoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6KZV
 
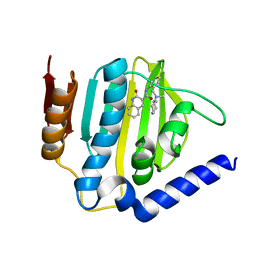 | |
6KZZ
 
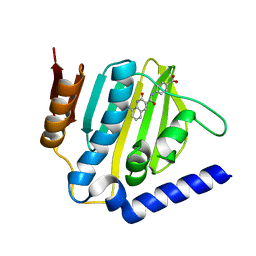 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
7C7N
 
 | |
7DEN
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-[(1~{R},5~{S})-6-[2-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]ethynyl]-3-azabicyclo[3.1.0]hexan-3-yl]butanoic acid, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Takashima, H. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7DEL
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 3-[4-[2-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenyl]propan-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Tanaka-Yamamoto, N. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7DEM
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 5-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]pent-4-yn-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Matsuda, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7CI5
 
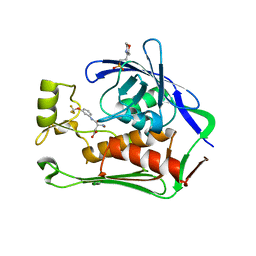 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (3R)-3-azanyl-4-oxidanylidene-4-[[3-(trifluoromethyloxy)phenyl]amino]butanoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI6
 
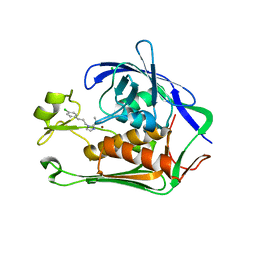 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[3-(4-chlorophenyl)propyl]imidazol-2-yl]ethanol, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI4
 
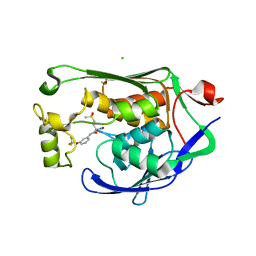 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-4-methylsulfonyl-N-[3-(trifluoromethyloxy)phenyl]butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI9
 
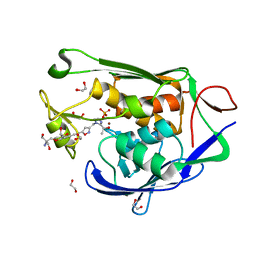 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-2-[[4-[2-[3-[[2-[(1S)-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenoxy]methyl]propane-1,3-diol, PHOSPHATE ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI7
 
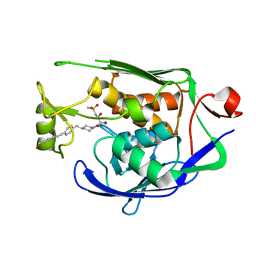 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R,3R)-2-azanyl-1-[4-[[4-[2-[4-(hydroxymethyl)phenyl]ethynyl]phenyl]methyl]piperidin-1-yl]-4-methylsulfonyl-3-oxidanyl-butan-1-one, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI8
 
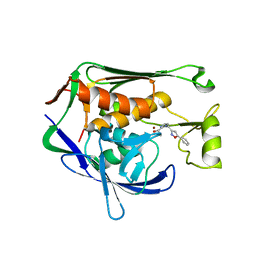 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[(5-phenyl-1,2-oxazol-3-yl)methyl]imidazol-2-yl]ethanol, MAGNESIUM ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
8K5W
 
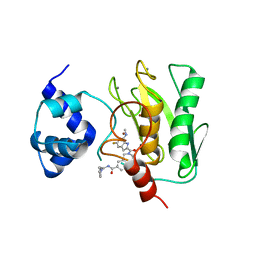 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8H78
 
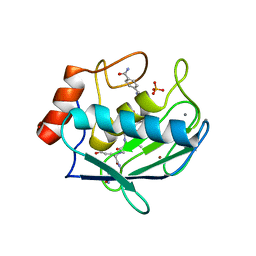 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | (2~{R})-2-[[4-[(4-aminocarbonylphenyl)carbonylamino]phenyl]sulfonylamino]-5-[(2~{S},4~{S})-4-azanyl-2-[[(2~{S})-1-[[(2~{S})-1-[(5-azanyl-5-oxidanylidene-pentyl)amino]-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamoyl]pyrrolidin-1-yl]-5-oxidanylidene-pentanoic acid, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Takeuchi, T, Mima, M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of TP0597850: A Selective, Chemically Stable, and Slow Tight-Binding Matrix Metalloproteinase-2 Inhibitor with a Phenylbenzamide-Pentapeptide Hybrid Scaffold.
J.Med.Chem., 66, 2023
|
|
7XJO
 
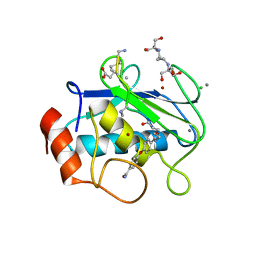 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Takeuchi, T. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aryloxyphenyl-Heptapeptide Hybrids as Potent and Selective Matrix Metalloproteinase-2 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 65, 2022
|
|
7XGJ
 
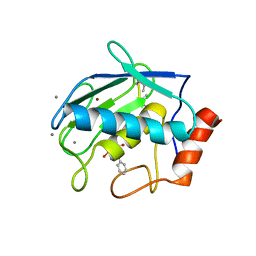 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | CALCIUM ION, GZS-ASN-ASP-ALA-LEU-IML-EOE-NH2, Matrix metalloproteinase-2, ... | | Authors: | Kamitani, M, Mima, M, Takeuchi, T. | | Deposit date: | 2022-04-05 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Aryloxyphenyl-Heptapeptide Hybrids as Potent and Selective Matrix Metalloproteinase-2 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 65, 2022
|
|
8K5Y
 
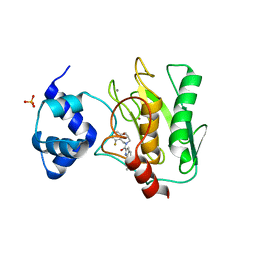 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
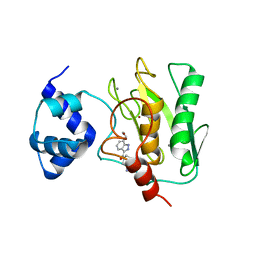 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
