8FGW
 
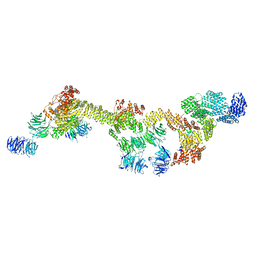 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
4PHM
 
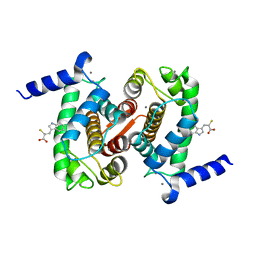 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | 3-(5-bromo-1H-indol-3-yl)-2-thioxopropanoic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
6URQ
 
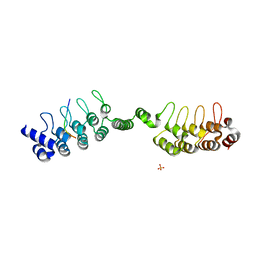 | | Complex structure of human poly-ADP-ribosyltransferase TNKS1 ARC2-ARC3 and P antigen family member 4 (PAGE4) | | Descriptor: | GLYCEROL, P antigen family member 4, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Zheng, Y, Koirala, S, Miller, D, Potts, P.R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tissue-Specific Regulation of the Wnt/ beta-Catenin Pathway by PAGE4 Inhibition of Tankyrase.
Cell Rep, 32, 2020
|
|
3H5N
 
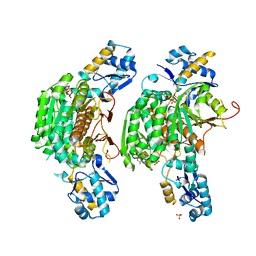 | | Crystal structure of E. coli MccB + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MccB protein, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
4PHK
 
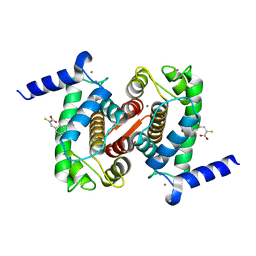 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids. The complex with (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid | | Descriptor: | (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B, Robinson, E. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4PHN
 
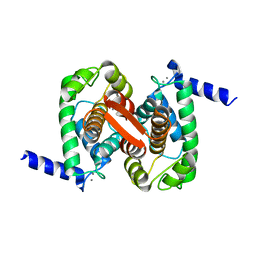 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | CALCIUM ION, Calpain small subunit 1 | | Authors: | Allemann, R.K, Rizkallah, P.J, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
3H5R
 
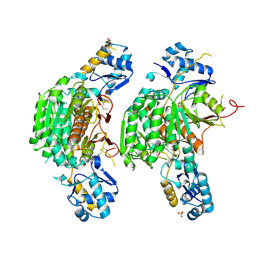 | | Crystal structure of E. coli MccB + Succinimide | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9G
 
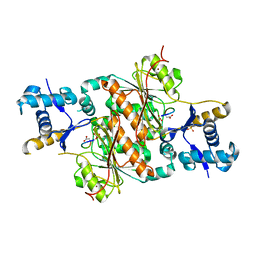 | | Crystal structure of E. coli MccB + MccA-N7isoASN | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H5A
 
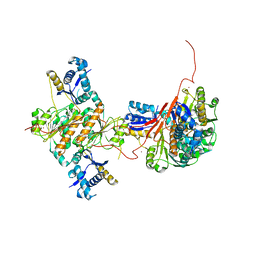 | | Crystal structure of E. coli MccB | | Descriptor: | MccB protein, ZINC ION | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9Q
 
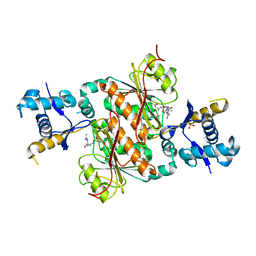 | | Crystal structure of E. coli MccB + SeMet MccA | | Descriptor: | MccB protein, Microcin C7 ANALOG, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9J
 
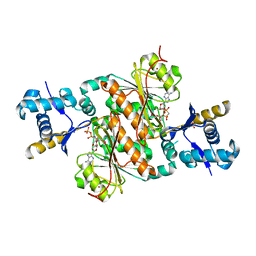 | | Crystal structure of E. coli MccB + AMPCPP + SeMeT MccA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MccB protein, Microcin C7 ANALOG, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
9BC9
 
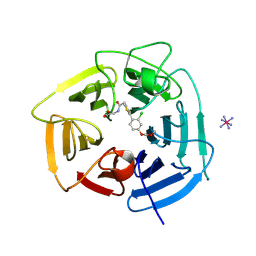 | | Structure of KLHDC2 bound to SJ10278 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[2-chloro-4-(methoxymethoxy)phenyl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
9BCC
 
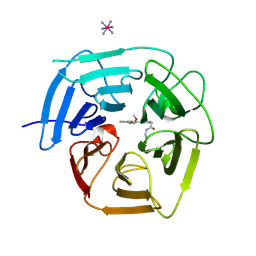 | | Structure of KLHDC2 bound to SJ46418 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
9BCA
 
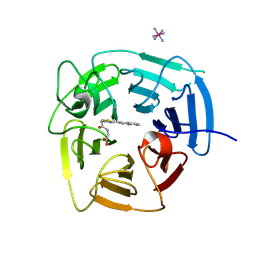 | | Structure of KLHDC2 bound to SJ46411 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-{[2-(naphthalen-2-yl)-1,3-thiazol-4-yl]acetyl}glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
7RYW
 
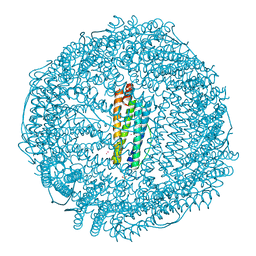 | | Crystal structure of Ferritin grown by the microbatch method in the presence of Agarose | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, D.J, Stojanoff, V. | | Deposit date: | 2021-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Ferritin grown by microbatch method in the presence of agarose
To Be Published
|
|
8FPE
 
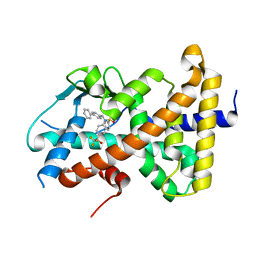 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | Descriptor: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2023-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4LCD
 
 | |
8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3IVQ
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-CiSBC2 | | Descriptor: | CiSBC2, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-PucSBC1_pep1 | | Descriptor: | PucSBC1, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
6AGK
 
 | | The structure of CH-II-77-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, H, Arnst, K, Wang, Y, Miller, D, Li, W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
4R32
 
 | | Crystal Structure Analysis of Pyk2 and Paxillin LD motifs | | Descriptor: | Paxillin, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.D, Nourse, A, Zheng, J.J. | | Deposit date: | 2014-08-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
3IVB
 
 | | Structures of SPOP-Substrate Complexes: Insights into Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-MacroH2ASBCpep1 | | Descriptor: | CHLORIDE ION, Core histone macro-H2A.1, Speckle-type POZ protein, ... | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure 9
To be Published
|
|
8F5Y
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
5C83
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 21 | | Descriptor: | (2R,5R)-4-[2-(6-benzyl-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
