3GEH
 
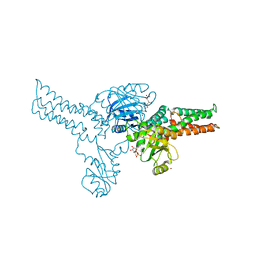 | | Crystal structure of MnmE from Nostoc in complex with GDP, FOLINIC ACID and ZN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ZINC ION, ... | | Authors: | Meyer, S, Wittinghofer, A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Kissing G domains of MnmE monitored by X-ray crystallography and pulse electron paramagnetic resonance spectroscopy
Plos Biol., 7, 2009
|
|
3GEE
 
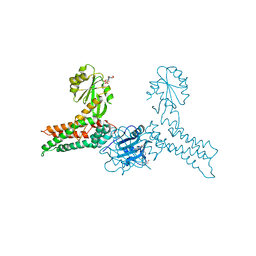 | | Crystal structure of MnmE from Chlorobium tepidum in complex with GDP and FOLINIC ACID | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, tRNA modification GTPase mnmE | | Authors: | Meyer, S, Wittinghofer, A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Kissing G domains of MnmE monitored by X-ray crystallography and pulse electron paramagnetic resonance spectroscopy
Plos Biol., 7, 2009
|
|
3GEI
 
 | |
3CP8
 
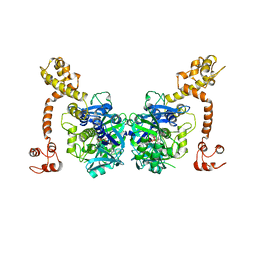 | | Crystal structure of GidA from Chlorobium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Meyer, S, Scrima, A, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
2JA3
 
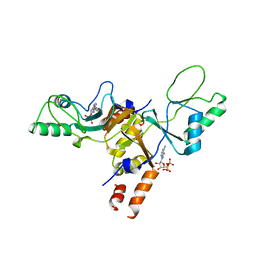 | | Cytoplasmic Domain of the Human Chloride Transporter ClC-5 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE CHANNEL PROTEIN 5 | | Authors: | Meyer, S, Savaresi, S, Forster, I.C, Dutzler, R. | | Deposit date: | 2006-11-21 | | Release date: | 2007-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nucleotide Recognition by the Cytoplasmic Domain of the Human Chloride Transporter Clc-5
Nat.Struct.Mol.Biol., 14, 2006
|
|
2J9L
 
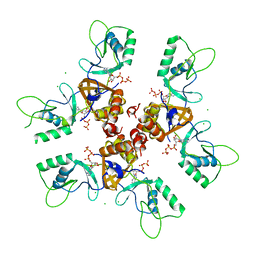 | | Cytoplasmic Domain of the Human Chloride Transporter ClC-5 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE CHANNEL PROTEIN 5, CHLORIDE ION | | Authors: | Meyer, S, Savaresi, S, Forster, I.C, Dutzler, R. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide Recognition by the Cytoplasmic Domain of the Human Chloride Transporter Clc-5
Nat.Struct.Mol.Biol., 14, 2006
|
|
3DA6
 
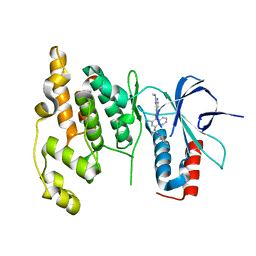 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EFW
 
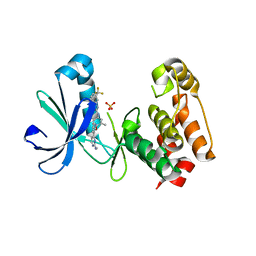 | | Structure of AuroraA with pyridyl-pyrimidine urea inhibitor | | Descriptor: | 1-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)phenyl]-3-[3-(trifluoromethyl)phenyl]urea, SULFATE ION, Serine/threonine-protein kinase 6 | | Authors: | Bellon, S.F, Cee, V, Hughes, P, Geuns-Meyer, S, Whittington, D. | | Deposit date: | 2008-09-10 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1R0K
 
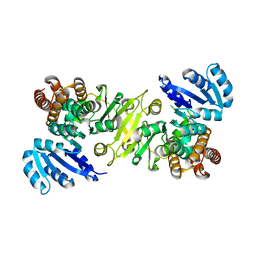 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Zymomonas mobilis | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ACETATE ION | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1R0L
 
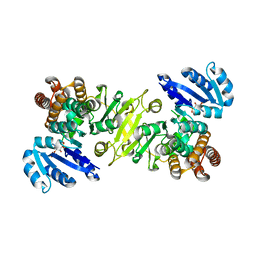 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from zymomonas mobilis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
3L8P
 
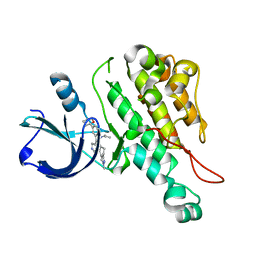 | | Crystal structure of cytoplasmic kinase domain of Tie2 complexed with inhibitor CEP11207 | | Descriptor: | 2-methyl-11-(1-methylethyl)-8-[(2S)-tetrahydro-2H-pyran-2-yl]-2,11,12,13-tetrahydro-4H-indazolo[5,4-a]pyrrolo[3,4-c]carbazol-4-one, Angiopoietin-1 receptor | | Authors: | Fedorov, A.A, Fedorov, E.V, Pauletti, D, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2010-01-03 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of cytoplasmic kinase domain of Tie2 complexed with inhibitor CEP11207
To be Published
|
|
3EG5
 
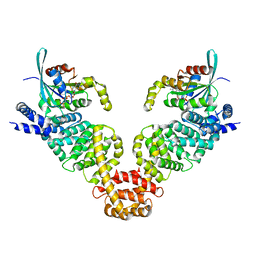 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
3F1J
 
 | | Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals RNA binding properties | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Matrix protein, SULFATE ION | | Authors: | Neumann, P, Lieber, D, Meyer, S, Dautel, P, Kerth, A, Kraus, I, Garten, W, Stubbs, M.T. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3CP2
 
 | | Crystal structure of GidA from E. coli | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Scrima, A, Meyer, S, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-30 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
4AQC
 
 | | Triazolopyridine-based Inhibitor of Janus Kinase 2 | | Descriptor: | 8-(4-methylsulfonylphenyl)-N-(4-morpholin-4-ylphenyl)-[1,2,4]triazolo[1,5-a]pyridin-2-amine, SULFATE ION, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Dugan, B.J, Gingrich, D.E, Mesaros, E.F, Milkiewicz, K.L, Curry, M.A, Zulli, A.L, Dobrzanski, P, Serdikoff, C, Jan, M, Angeles, T.S, Albom, M.S, Mason, J.L, Aimone, L.D, Meyer, S.L, Huang, Z, Wells-Knecht, K.J, Ator, M.A, Ruggeri, B.A, Dorsey, B.D. | | Deposit date: | 2012-04-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Selective, Orally Bioavailable 1,2,4-Triazolo[1,5-A]Pyridine-Based Inhibitor of Janus Kinase 2 for Use in Anticancer Therapy: Discovery of Cep-33779.
J.Med.Chem., 55, 2012
|
|
2D4Z
 
 | |
3DTC
 
 | | Crystal structure of mixed-lineage kinase MLK1 complexed with compound 16 | | Descriptor: | 12-(2-hydroxyethyl)-2-(1-methylethoxy)-13,14-dihydronaphtho[2,1-a]pyrrolo[3,4-c]carbazol-5(12H)-one, Mitogen-activated protein kinase kinase kinase 9, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2008-07-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mixed-lineage kinase 1 and mixed-lineage kinase 3 subtype-selective dihydronaphthyl[3,4-a]pyrrolo[3,4-c]carbazole-5-ones: optimization, mixed-lineage kinase 1 crystallography, and oral in vivo activity in 1-methyl-4-phenyltetrahydropyridine models.
J.Med.Chem., 51, 2008
|
|
2BJU
 
 | | Plasmepsin II complexed with a highly active achiral inhibitor | | Descriptor: | N-(R-CARBOXY-ETHYL)-ALPHA-(S)-(2-PHENYLETHYL), PLASMEPSIN II | | Authors: | Prade, L, Jones, A.F, Boss, C, Richards-Bildstein, S, Meyer, S, Binkert, C, Bur, D. | | Deposit date: | 2005-02-08 | | Release date: | 2005-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-Ray Structure of Plasmepsin II Complexed with a Potent Achiral Inhibitor.
J.Biol.Chem., 280, 2005
|
|
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human dopamine transporter in complex with cocaine.
Nature, 632, 2024
|
|
1K9V
 
 | | Structural evidence for ammonia tunelling across the (beta-alpha)8-barrel of the imidazole glycerol phosphate synthase bienzyme complex | | Descriptor: | ACETIC ACID, Amidotransferase hisH | | Authors: | Douangamath, A, Walker, M, Beismann-Driemeyer, S, Vega-Fernandez, M.C, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-10-31 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
Structure, 10, 2002
|
|
1GPW
 
 | | Structural evidence for ammonia tunneling across the (beta/alpha)8 barrel of the imidazole glycerol phosphate synthase bienzyme complex. | | Descriptor: | AMIDOTRANSFERASE HISH, HISF PROTEIN, PHOSPHATE ION | | Authors: | Walker, M, Beismann-Driemeyer, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Evidence for Ammonia Tunneling Across the (Beta Alpha)(8) Barrel of the Imidazole Glycerol Phosphate Synthase Bienzyme Complex.
Structure, 10, 2002
|
|
1IRA
 
 | | COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERLEUKIN-1 RECEPTOR, INTERLEUKIN-1 RECEPTOR ANTAGONIST | | Authors: | Schreuder, H.A, Tardif, C, Tramp-Kalmeyer, S, Soffientini, A, Sarubbi, E, Akeson, A, Bowlin, T, Yanofsky, S, Barrett, R.W. | | Deposit date: | 1998-04-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist.
Nature, 386, 1997
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
1DDB
 
 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (BID) | | Authors: | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | Deposit date: | 1999-02-19 | | Release date: | 1999-08-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
