1CNT
 
 | | CILIARY NEUROTROPHIC FACTOR | | Descriptor: | CILIARY NEUROTROPHIC FACTOR, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Mcdonald, N.Q, Panayotatos, N, Hendrickson, W.A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-03-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric human ciliary neurotrophic factor determined by MAD phasing.
EMBO J., 14, 1995
|
|
6FEK
 
 | |
1BET
 
 | | NEW PROTEIN FOLD REVEALED BY A 2.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF NERVE GROWTH FACTOR | | Descriptor: | BETA-NERVE GROWTH FACTOR | | Authors: | Mcdonald, N.Q, Lapatto, R, Murray-Rust, J, Gunning, J, Wlodawer, A, Blundell, T.L. | | Deposit date: | 1993-04-08 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor.
Nature, 354, 1991
|
|
8OS6
 
 | | Structure of a GFRA1/GDNF LICAM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houghton, F.M, Adams, S.E, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Architecture and regulation of a GDNF-GFR alpha 1 synaptic adhesion assembly.
Nat Commun, 14, 2023
|
|
1SGF
 
 | | CRYSTAL STRUCTURE OF 7S NGF: A COMPLEX OF NERVE GROWTH FACTOR WITH FOUR BINDING PROTEINS (SERINE PROTEINASES) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NERVE GROWTH FACTOR, ... | | Authors: | Bax, B.D.V, Blundell, T.L, Murray-Rust, J, Mcdonald, N.Q. | | Deposit date: | 1997-08-08 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins.
Structure, 5, 1997
|
|
7NZN
 
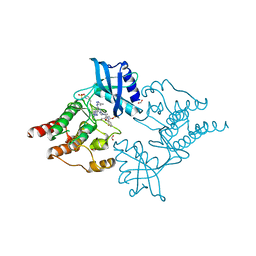 | | Structure of RET kinase domain bound to inhibitor JB-48 | | Descriptor: | 2-[4-[[4-[1-[2-(dimethylamino)ethyl]pyrazol-4-yl]-6-[(3-methyl-1~{H}-pyrazol-5-yl)amino]pyrimidin-2-yl]amino]phenyl]-~{N}-(3-fluorophenyl)ethanamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2021-03-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-Trisubstituted Pyrimidine Derivatives as Type I RET and RET Gatekeeper Mutant Inhibitors with a Novel Kinase Binding Pose.
J.Med.Chem., 65, 2022
|
|
4UU6
 
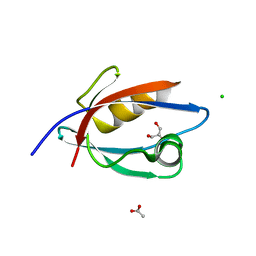 | | CRYSTAL STRUCTURE OF THE PDZ DOMAIN OF PALS1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5AMN
 
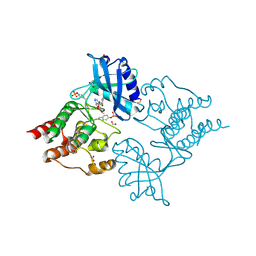 | | The Discovery of 2-Substituted Phenol Quinazolines as Potent and Selective RET Kinase Inhibitors | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Newton, R, Bowler, K, Burns, E.M, Chapman, P, Fairweather, E, Fritzl, S, Goldberg, K, Hamilton, N.M, Holt, S.V, Hopkins, G.V, Jones, S.D, Jordan, A.M, Lyons, A, McDonald, N.Q, Maguire, L.A, Mould, D.P, Purkiss, A.G, Small, H.F, Stowell, A, Thomson, G.J, Waddell, I.D, Waszkowycz, B, Watson, A.J, Ogilvie, D.J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The discovery of 2-substituted phenol quinazolines as potent RET kinase inhibitors with improved KDR selectivity.
Eur J Med Chem, 112, 2016
|
|
6SXB
 
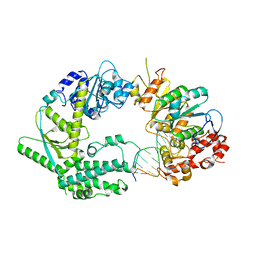 | | XPF-ERCC1 Cryo-EM Structure, DNA-Bound form | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*AP*TP*GP*CP*TP*GP*A)-3'), DNA excision repair protein ERCC-1, ... | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
6SXA
 
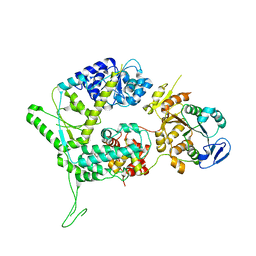 | | XPF-ERCC1 Cryo-EM Structure, Apo-form | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
5FM2
 
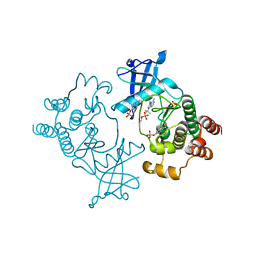 | | Crystal structure of hyper-phosphorylated RET kinase domain with (proximal) juxtamembrane segment | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Barry, R, Borg, A, Orme, M, Mouilleron, S, Martinez-Torres, R.J, Meier, P, McDonald, N.Q. | | Deposit date: | 2015-10-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RET Functions as a Dual-Specificity Kinase that Requires Allosteric Inputs from Juxtamembrane Elements.
Cell Rep, 17, 2016
|
|
1UW5
 
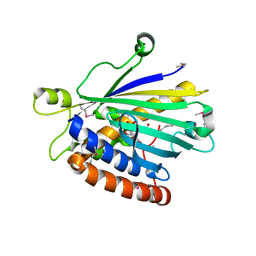 | | Structure of PITP-alpha complexed to phosphatidylinositol | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, PHOSPHATIDYLINOSITOL TRANSFER PROTEIN ALPHA ISOFORM | | Authors: | Tilley, S.J, Skippen, A, Murray-Rust, J, Cockcroft, S, McDonald, N.Q. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Function Analysis of Human [Corrected] Phosphatidylinositol Transfer Protein Alpha Bound to Phosphatidylinositol.
Structure, 12, 2004
|
|
5FM3
 
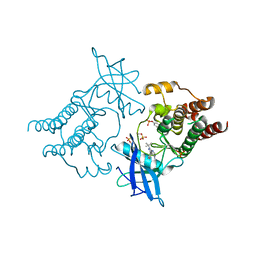 | | Crystal structure of hyper-phosphorylated RET kinase domain with (proximal) juxtamembrane segment | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Barry, R, Borg, A, Orme, M, Mouilleron, S, Martinez-Torres, R.J, Meier, P, McDonald, N.Q. | | Deposit date: | 2015-10-30 | | Release date: | 2016-12-28 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | RET Functions as a Dual-Specificity Kinase that Requires Allosteric Inputs from Juxtamembrane Elements.
Cell Rep, 17, 2016
|
|
7AML
 
 | | RET/GDNF/GFRa1 extracellular complex Cryo-EM structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-10-09 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7AMK
 
 | | Zebrafish RET Cadherin Like Domains 1 to 4. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Purkiss, A.G, McDonald, N.Q, Goodman, K.M, Narowtek, A, Knowles, P.P. | | Deposit date: | 2020-10-09 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7AB8
 
 | | Crystal structure of a GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
1MKP
 
 | | CRYSTAL STRUCTURE OF PYST1 (MKP3) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PYST1 | | Authors: | Stewart, A.E, Dowd, S, Keyse, S, Mcdonald, N.Q. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation.
Nat.Struct.Biol., 6, 1999
|
|
1E3Y
 
 | |
1E41
 
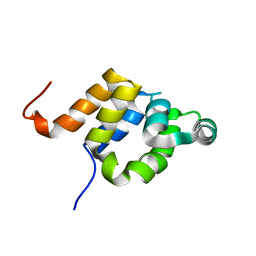 | |
1F5Q
 
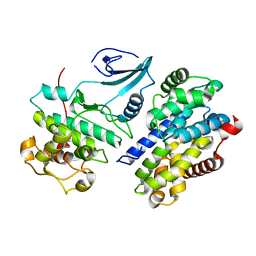 | | CRYSTAL STRUCTURE OF MURINE GAMMA HERPESVIRUS CYCLIN COMPLEXED TO HUMAN CYCLIN DEPENDENT KINASE 2 | | Descriptor: | CHLORIDE ION, CYCLIN DEPENDENT KINASE 2, GAMMA HERPESVIRUS CYCLIN | | Authors: | Card, G.L, Knowles, P, Laman, H, Jones, N, McDonald, N.Q. | | Deposit date: | 2000-06-15 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a gamma-herpesvirus cyclin-cdk complex.
EMBO J., 19, 2000
|
|
1GR0
 
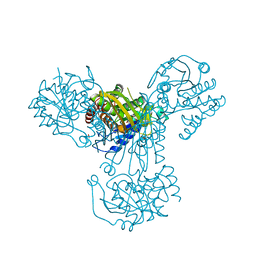 | | myo-inositol 1-phosphate synthase from Mycobacterium tuberculosis in complex with NAD and zinc. | | Descriptor: | CACODYLATE ION, INOSITOL-3-PHOSPHATE SYNTHASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Norman, R.A, Murray-Rust, J, McDonald, N.Q, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Inositol 1-Phosphate Synthase from Mycobacterium Tuberculosis, a Key Enzyme in Phosphatidylinositol Synthesis
Structure, 10, 2002
|
|
4UX8
 
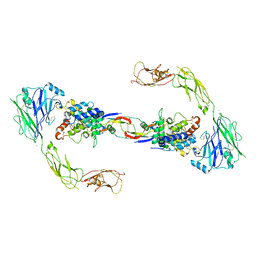 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
4UU5
 
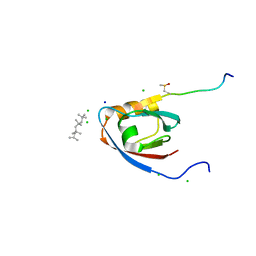 | | CRYSTAL STRUCTURE OF THE PDZ DOMAIN OF PALS1 IN COMPLEX WITH THE CRB PEPTIDE | | Descriptor: | 2,2,4,4,6,6,8-heptamethylnonane, CHLORIDE ION, MAGUK P55 SUBFAMILY MEMBER 5, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5LIH
 
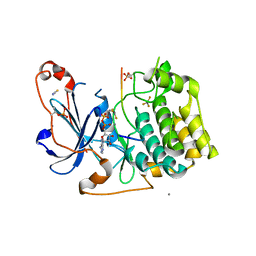 | | Structure of a peptide-substrate bound to PKCiota core kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MANGANESE (II) ION, ... | | Authors: | Soriano, E.V, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
5LI9
 
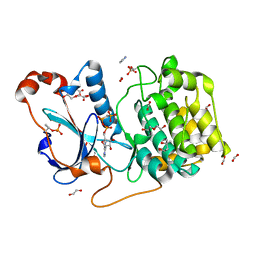 | | Structure of a nucleotide-bound form of PKCiota core kinase domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
