8C3W
 
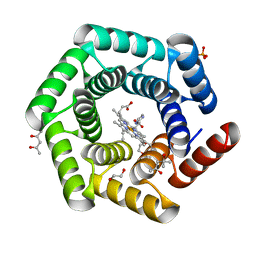 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ortmayer, M, Levy, C. | | 登録日 | 2022-12-29 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
6Y1T
 
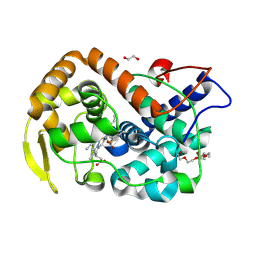 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Ortmayer, M, Levy, C, Green, A.P. | | 登録日 | 2020-02-13 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6Y2Y
 
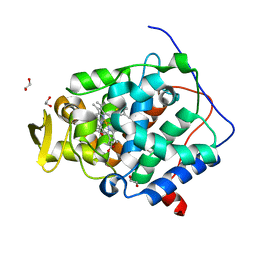 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with Trp51 to S-Trp51 and Trp191Phe modifications | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Ortmayer, M, Levy, C, Green, A.P. | | 登録日 | 2020-02-17 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
5LTE
 
 | |
5LTI
 
 | |
5LTH
 
 | |
6H08
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a His175Me-His proximal ligand substitution | | 分子名称: | COBALT (II) ION, Cytochrome c peroxidase, mitochondrial, ... | | 著者 | Ortmayer, M, Levy, C, Green, A.P. | | 登録日 | 2018-07-06 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rewiring the "Push-Pull" Catalytic Machinery of a Heme Enzyme Using an Expanded Genetic Code.
Acs Catalysis, 10, 2020
|
|
6Z0P
 
 | | BceF Tyrosine Kinase Domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BceF | | 著者 | Landau, M, Mayer, M, Abd Alhadi, M, Dvir, H. | | 登録日 | 2020-05-10 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Functional Insights into the Biofilm-Associated BceF Tyrosine Kinase Domain from Burkholderia cepacia .
Biomolecules, 11, 2021
|
|
5M39
 
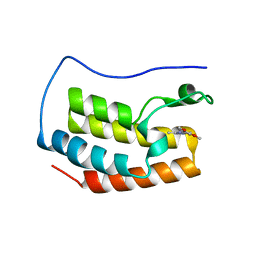 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 1 | | 分子名称: | 6-(3,4-dimethoxyphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | 著者 | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | 登録日 | 2016-10-14 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5M3A
 
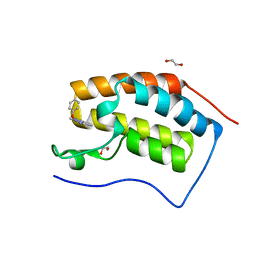 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | 分子名称: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | 著者 | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | 登録日 | 2016-10-14 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
1LB8
 
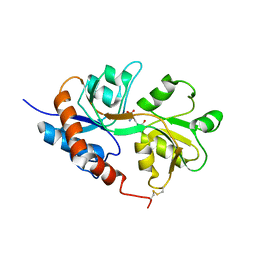 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | 分子名称: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | 著者 | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | 登録日 | 2002-04-02 | | 公開日 | 2002-06-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LB9
 
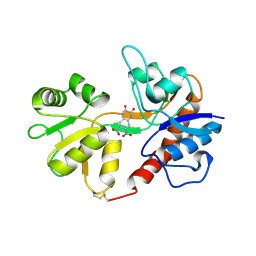 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with antagonist DNQX at 2.3 A resolution | | 分子名称: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | 著者 | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | 登録日 | 2002-04-02 | | 公開日 | 2002-06-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBB
 
 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | 著者 | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | 登録日 | 2002-04-02 | | 公開日 | 2002-06-05 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBC
 
 | | Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution | | 分子名称: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamine Receptor 2, ... | | 著者 | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | 登録日 | 2002-04-02 | | 公開日 | 2002-05-29 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
8PIF
 
 | |
6BIT
 
 | | SIRPalpha antibody complex | | 分子名称: | KWAR23 Fab heavy chain, KWAR23 Fab light chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Ring, N.G, Herndler-Brandstetter, D, Weiskopf, K, Shan, L, Volkmer, J.P, George, B.M, Lietzenmayer, M, McKenna, K.M, Naik, T.J, McCarty, A, Zheng, Y, Ring, A.M, Flavell, R.A, Weissman, I.L. | | 登録日 | 2017-11-03 | | 公開日 | 2017-12-06 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.191 Å) | | 主引用文献 | Anti-SIRP alpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5A3B
 
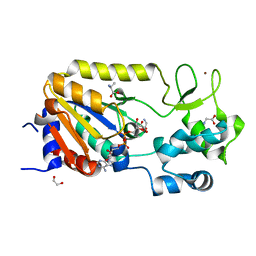 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with ADP-ribose | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, ALANINE, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3A
 
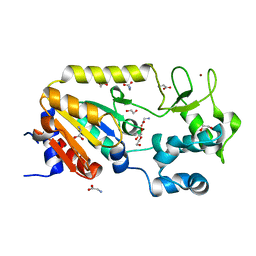 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | 分子名称: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
1V9U
 
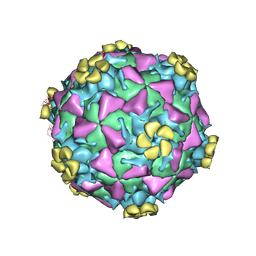 | | Human Rhinovirus 2 bound to a fragment of its cellular receptor protein | | 分子名称: | CALCIUM ION, Coat protein VP1, Coat protein VP2, ... | | 著者 | Verdaguer, N, Fita, I, Reithmayer, M, Moser, R, Blaas, D. | | 登録日 | 2004-02-03 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
5A35
 
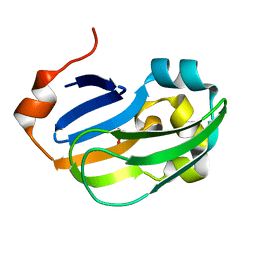 | | Crystal structure of Glycine Cleavage Protein H-Like (GcvH-L) from Streptococcus pyogenes | | 分子名称: | GLYCINE CLEAVAGE SYSTEM H PROTEIN, PENTAETHYLENE GLYCOL | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-27 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3C
 
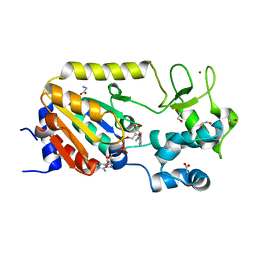 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | 分子名称: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
