6ORR
 
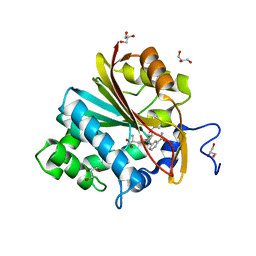 | | Co-crystal structure of human NicotinamideN-Methyltransferase (NNMT) in complex with High-Affinity Alkynyl Bisubstrate Inhibitor NS1 | | Descriptor: | 9-{9-amino-6-[(3-carbamoylphenyl)ethynyl]-5,6,7,8,9-pentadeoxy-D-glycero-alpha-L-talo-decofuranuronosyl}-9H-purin-6-amine, GLYCEROL, NNMT protein | | Authors: | May, E.J, Policarpo, R.L, Gaudet, R. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-Affinity Alkynyl Bisubstrate Inhibitors of NicotinamideN-Methyltransferase (NNMT).
J.Med.Chem., 62, 2019
|
|
6E7L
 
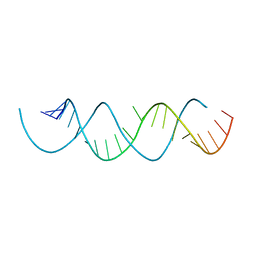 | | Structure of a dimerized UUCG motif | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*UP*GP*CP*CP*C)-3') | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2018-07-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of an RNA helix with pyrimidine mismatches and cross-strand stacking.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6PPN
 
 | | Structure of S. pombe Lsm2-8 with unprocessed U6 snRNA | | Descriptor: | Mimic of unprocessed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
6PPQ
 
 | |
6PPV
 
 | |
6PPP
 
 | | Structure of S. pombe Lsm2-8 with processed U6 snRNA | | Descriptor: | Mimic of processed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
5V1M
 
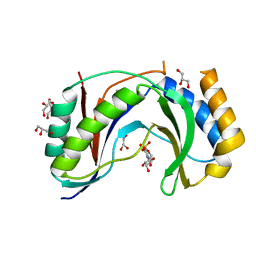 | | Structure of human Usb1 with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, U6 snRNA phosphodiesterase, ... | | Authors: | Montemayor, E.J, Didychuk, A.L, Butcher, S.E. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
5VSU
 
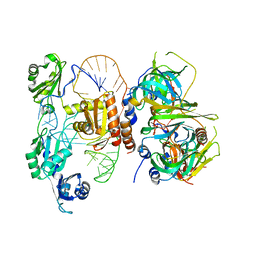 | | Structure of yeast U6 snRNP with 2'-phosphate terminated U6 RNA | | Descriptor: | Saccharomyces cerevisiae strain T8 chromosome XII sequence, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA-associated Sm-like protein LSm2, ... | | Authors: | Montemayor, E.J. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Architecture of the U6 snRNP reveals specific recognition of 3'-end processed U6 snRNA.
Nat Commun, 9, 2018
|
|
5TF6
 
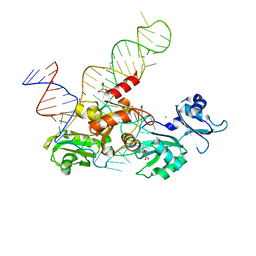 | | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Montemayor, E.J, Brow, D.A, Butcher, S.E. | | Deposit date: | 2016-09-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4PEF
 
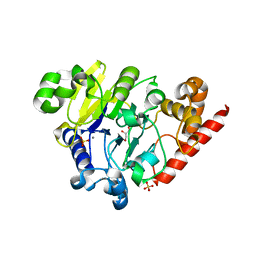 | | Dbr1 in complex with sulfate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA lariat debranching enzyme, ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PEI
 
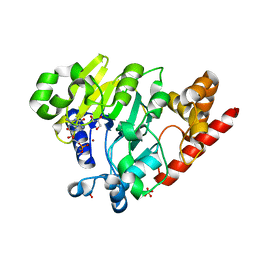 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PEG
 
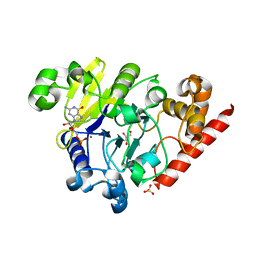 | | Dbr1 in complex with guanosine-5'-monophosphate | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4N0T
 
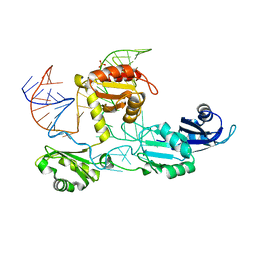 | | Core structure of the U6 small nuclear ribonucleoprotein at 1.7 Angstrom resolution | | Descriptor: | SULFATE ION, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA | | Authors: | Montemayor, E.J, Curran, E.C, Liao, H, Andrews, K.L, Treba, C.N, Butcher, S.E, Brow, D.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Core structure of the U6 small nuclear ribonucleoprotein at 1.7- angstrom resolution.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PEH
 
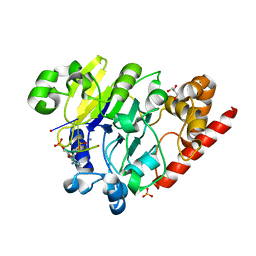 | | Dbr1 in complex with synthetic linear RNA | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA (5'-R(*CP*UP*AP*(A2P)P*AP*CP*AP*A)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
3BJ8
 
 | |
3BJ7
 
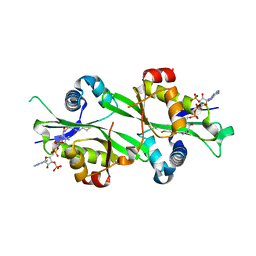 | |
2XCT
 
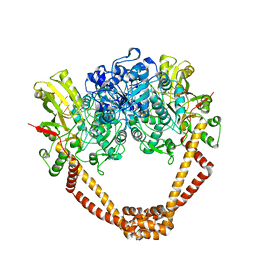 | | The twinned 3.35A structure of S. aureus Gyrase complex with Ciprofloxacin and DNA | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 5'-D(AP*GP*CP*CP*GP*TP*AP*G)-3', 5'-D(GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3', ... | | Authors: | Bax, B.D, Chan, P, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCR
 
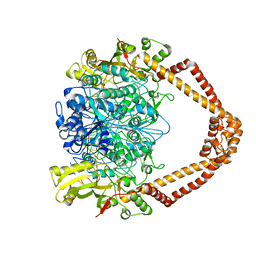 | | The 3.5A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase complexed with GSK299423 and DNA | | Descriptor: | 5'-D(*5UA*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCS
 
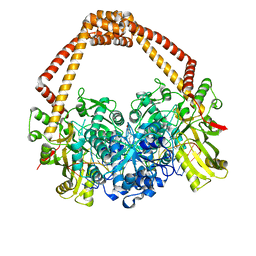 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCQ
 
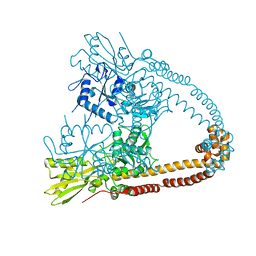 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCO
 
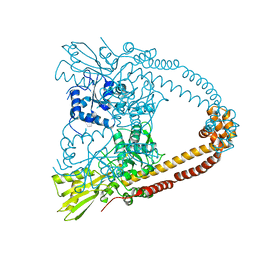 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
1F1Z
 
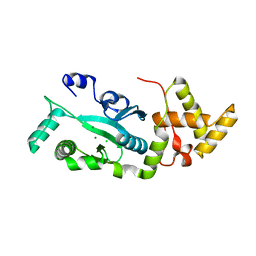 | | TNSA, a catalytic component of the TN7 transposition system | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TNSA ENDONUCLEASE | | Authors: | Hickman, A.B, Li, Y, Mathew, S.V, May, E.W, Craig, N.L, Dyda, F. | | Deposit date: | 2000-05-21 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unexpected structural diversity in DNA recombination: the restriction endonuclease connection.
Mol.Cell, 5, 2000
|
|
9CEP
 
 | | Caulobacter crescentus FljJN flagellar filament (asymmetrical) | | Descriptor: | Flagellin | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2024-06-26 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
9CEF
 
 | | Caulobacter crescentus FljN flagellar filament (symmetrized) | | Descriptor: | Flagellin FljN | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2024-06-26 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
9CEO
 
 | | Caulobacter crescentus FljJM flagellar filament (asymmetrical) | | Descriptor: | Flagellin FljM | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2024-06-26 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
