7RY1
 
 | | human Hsp90_MC domain structure | | 分子名称: | Heat shock protein HSP 90-alpha, N-[2-(1-MALEIMIDYL)ETHYL]-7-DIETHYLAMINOCOUMARIN-3-CARBOXAMIDE | | 著者 | Peng, S, Deng, J, Matts, R. | | 登録日 | 2021-08-24 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.523 Å) | | 主引用文献 | Crystal structure of the middle and C-terminal domains of Hsp90 alpha labeled with a coumarin derivative reveals a potential allosteric binding site as a drug target.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5UCJ
 
 | | Hsp90b N-terminal domain with inhibitors | | 分子名称: | (5-fluoroisoindolin-2-yl)(4-hydroxy-5-isopropylbenzo[d]isoxazol-7-yl)methanone, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-beta | | 著者 | Peng, S, Balch, M, Matts, R, Deng, J. | | 登録日 | 2016-12-22 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.693 Å) | | 主引用文献 | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
5UCI
 
 | | Hsp90b N-terminal domain with inhibitors | | 分子名称: | (2,4-Dihydroxy-3-(hydroxymethyl)-5-isopropylphenyl)(isoindolin-2-yl)methanone, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Peng, S, Balch, M, Matts, R, Deng, J. | | 登録日 | 2016-12-22 | | 公開日 | 2018-01-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
5UCH
 
 | | Hsp90b N-terminal domain with inhibitors | | 分子名称: | 2-(5-Hydroxy-4-(isoindoline-2-carbonyl)-2-isopropylphenyl)acetonitrile, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Peng, S, Balch, M, Matts, R, Deng, J. | | 登録日 | 2016-12-22 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.654 Å) | | 主引用文献 | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
5UC4
 
 | | Hsp90b N-terminal domain with inhibitors | | 分子名称: | 5-Hydroxy-4-(isoindoline-2-carbonyl)-2-isopropylbenzaldehyde, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Deng, J, Peng, S, Balch, M, Matts, R. | | 登録日 | 2016-12-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
7LSZ
 
 | | Hsp90a N-terminal inhibitor | | 分子名称: | Heat shock protein HSP 90-alpha, {3-[(2,3-dihydro-1,4-benzodioxin-6-yl)sulfanyl]-4-hydroxyphenyl}(1,3-dihydro-2H-isoindol-2-yl)methanone | | 著者 | Balch, M, Peng, S, Deng, J, Matts, R. | | 登録日 | 2021-02-18 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LT0
 
 | | Hsp90a N-terminal inhibitor | | 分子名称: | (1,3-dihydro-2H-isoindol-2-yl){3-[(3,4-dimethylphenyl)sulfanyl]-4-hydroxyphenyl}methanone, Heat shock protein HSP 90-alpha | | 著者 | Balch, M, Peng, S, Deng, J, Matts, R. | | 登録日 | 2021-02-18 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7RY0
 
 | | human Hsp90_MC domain structure | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, Heat shock protein HSP 90-alpha | | 著者 | Peng, S, Deng, J, Matts, R. | | 登録日 | 2021-08-24 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
7UR3
 
 | | Hsp90 alpha inhibitor | | 分子名称: | (5-fluoro-1,3-dihydro-2H-isoindol-2-yl){4-hydroxy-3-[(2S)-2-hydroxy-5-phenylpentan-2-yl]phenyl}methanone, Heat shock protein HSP 90-alpha | | 著者 | Deng, J, Peng, S, Balch, M, Matts, R. | | 登録日 | 2022-04-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Activity Relationship Study of Tertiary Alcohol Hsp90 alpha-Selective Inhibitors with Novel Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
7RXZ
 
 | | human Hsp90_MC domain structure | | 分子名称: | Heat shock protein HSP 90-alpha | | 著者 | Peng, S, Deng, J, Matts, R. | | 登録日 | 2021-08-24 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.152 Å) | | 主引用文献 | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
7U8W
 
 | | hTRAP1 with inhibitors | | 分子名称: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-fluoro-2H-isoindol-2-yl)methanone | | 著者 | Deng, J, Matts, R, Peng, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.706 Å) | | 主引用文献 | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8X
 
 | | hTRAP1 with inhibitors | | 分子名称: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-methoxy-2H-isoindol-2-yl)methanone | | 著者 | Deng, J, Matts, R, Peng, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8V
 
 | | hTRAP1 with inhibitors | | 分子名称: | (4-hydroxy-1,3-phenylene)bis[(2H-isoindol-2-yl)methanone], Heat shock protein 75 kDa, mitochondrial | | 著者 | Deng, J, Matts, R, Peng, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.448 Å) | | 主引用文献 | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8U
 
 | | hTRAP1 with inhibitors | | 分子名称: | Heat shock protein 75 kDa, mitochondrial, [2-hydroxy-5-(2H-isoindole-2-carbonyl)phenyl]{5-[3-(triphenyl-lambda~5~-phosphanyl)propoxy]-2H-isoindol-2-yl}methanone | | 著者 | Deng, J, Matts, R, Peng, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.065 Å) | | 主引用文献 | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
4FNT
 
 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose | | 分子名称: | Alpha-galactosidase AgaA, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | 著者 | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | 登録日 | 2012-06-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FNP
 
 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus | | 分子名称: | Alpha-galactosidase AgaA, SULFATE ION | | 著者 | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | 登録日 | 2012-06-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FNQ
 
 | | Crystal structure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus | | 分子名称: | 1,2-ETHANEDIOL, Alpha-galactosidase AgaB | | 著者 | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | 登録日 | 2012-06-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FNU
 
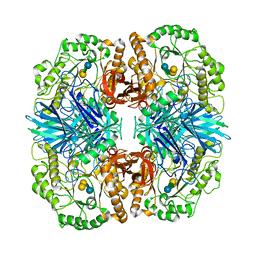 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose | | 分子名称: | Alpha-galactosidase AgaA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose | | 著者 | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | 登録日 | 2012-06-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FNR
 
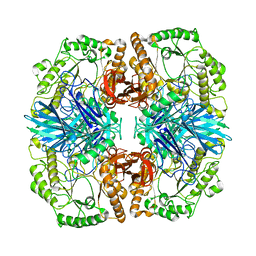 | | Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus | | 分子名称: | Alpha-galactosidase AgaA | | 著者 | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | 登録日 | 2012-06-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FNS
 
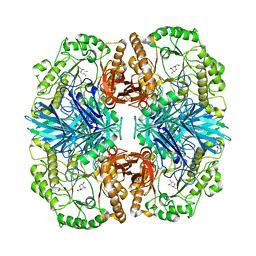 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin | | 分子名称: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-galactosidase AgaA, ... | | 著者 | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | 登録日 | 2012-06-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4U15
 
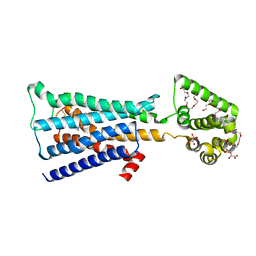 | | M3-mT4L receptor bound to tiotropium | | 分子名称: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, D(-)-TARTARIC ACID, ... | | 著者 | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | 登録日 | 2014-07-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
4U16
 
 | | M3-mT4L receptor bound to NMS | | 分子名称: | D(-)-TARTARIC ACID, Muscarinic acetylcholine receptor M3,Lysozyme,Muscarinic acetylcholine receptor M3, N-methyl scopolamine | | 著者 | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | 登録日 | 2014-07-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
