1BX4
 
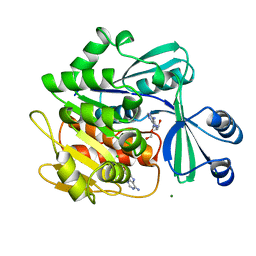 | | STRUCTURE OF HUMAN ADENOSINE KINASE AT 1.50 ANGSTROMS | | Descriptor: | ADENOSINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mathews, I.I, Erion, M.D, Ealick, S.E. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of human adenosine kinase at 1.5 A resolution.
Biochemistry, 37, 1998
|
|
1FPC
 
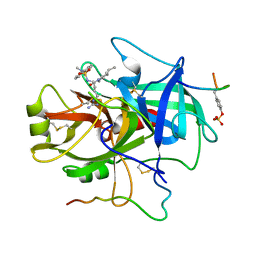 | | ACTIVE SITE MIMETIC INHIBITION OF THROMBIN | | Descriptor: | Hirudin, amino{[(4S)-4-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)-5-(4-ethylpiperidin-1-yl)-5-oxopentyl]amino}methaniminium, thrombin | | Authors: | Tulinsky, A, Mathews, I.I. | | Deposit date: | 1994-10-16 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site mimetic inhibition of thrombin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
8VTO
 
 | |
7SH1
 
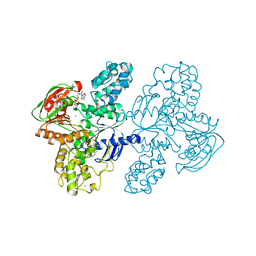 | | Class II UvrA protein - Ecm16 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Excinuclease ABC subunit UvrA, ... | | Authors: | Grade, P, Erlandson, A, Ullah, A, Mathews, I.I, Chen, X, Kim, C.-Y, Mera, P.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.
Sci Rep, 13, 2023
|
|
8T3P
 
 | |
8VTL
 
 | |
8VTM
 
 | |
8VTN
 
 | |
8VTK
 
 | |
4D8P
 
 | | Structural and functional studies of the trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) molecule | | Descriptor: | GLYCEROL, HLA-DQA1 protein, Peptide from Gamma-gliadin,HLA class II histocompatibility antigen, ... | | Authors: | Kim, C.-Y, Hotta, K, Mathews, I.I, Chen, X. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule
J.Biol.Chem., 287, 2012
|
|
2ATE
 
 | | Structure of the complex of PurE with NitroAIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kappock, T.J, Mathews, I.I, Zaugg, J.B, Peng, P, Hoskins, A.A, Okamoto, A, Ealick, S.E, Stubbe, J. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
Biochemistry, 46, 2007
|
|
1HLT
 
 | |
2BKL
 
 | | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Inter-Domain Dynamics in Catalysis and Specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, N-[(BENZYLOXY)CARBONYL]-L-ALANYL-L-PROLINE, PROLYL ENDOPEPTIDASE, ... | | Authors: | Khosla, C, Shan, L, Mathews, I.I. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7T2U
 
 | |
7SQL
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with a weak small molecule inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-(4-bromophenyl)-2-{[1-(4-fluorophenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]sulfanyl}acetamide, ... | | Authors: | Mashayekh, S, Stunkard, L.M, Kienle, M, Mathews, I.I, Khosla, C. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prototyping of Allosteric Inhibitors of Human Uridine/Cytidine Kinase 2 (UCK2).
Biochemistry, 61, 2022
|
|
7U6Q
 
 | | TEM-1 beta-lactamase | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-03-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
7MHA
 
 | |
3HAT
 
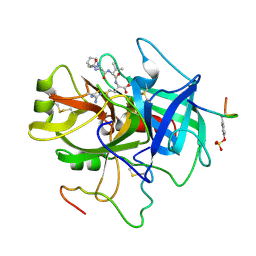 | | ACTIVE SITE MIMETIC INHIBITION OF THROMBIN | | Descriptor: | FPAM (FIBRINOPEPTIDE A MIMIC), Hirudin variant-2, Thrombin heavy chain, ... | | Authors: | Tulinsky, A, Mathews, I.I. | | Deposit date: | 1994-10-16 | | Release date: | 1995-02-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active-site mimetic inhibition of thrombin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
6C9U
 
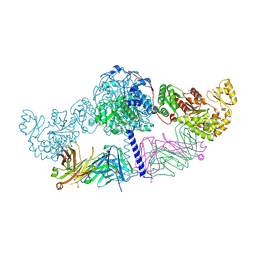 | | Crystal structure of [KS3][AT3] didomain from module 3 of 6-deoxyerthronolide B synthase in complex with antibody fragment (Fab) | | Descriptor: | 6-deoxyerythronolide-B synthase EryA2, modules 3 and 4, Heavy chain of Fab 1B2, ... | | Authors: | Deis, L.N, Li, X, Mathews, I.I, Khosla, C. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Function Analysis of the Extended Conformation of a Polyketide Synthase Module.
J. Am. Chem. Soc., 140, 2018
|
|
4RA0
 
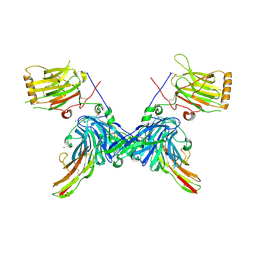 | | An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kariolis, M.S, Kapur, S, Mathews, I.I, Cochran, J.R. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis.
Nat.Chem.Biol., 10, 2014
|
|
4RDS
 
 | |
6APM
 
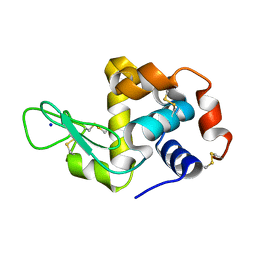 | | Hen egg-white lysozyme (WT), solved with serial millisecond crystallography using synchrotron radiation | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
6APK
 
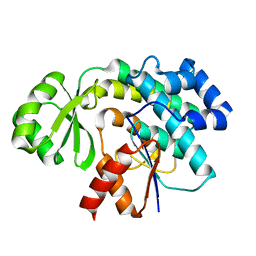 | | Trans-acting transferase from Disorazole synthase solved by serial femtosecond XFEL crystallography | | Descriptor: | DisD protein | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Khosla, C, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
6WH9
 
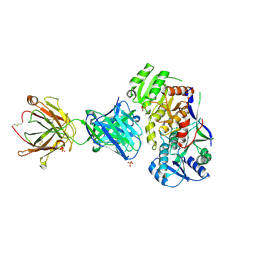 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
6V2J
 
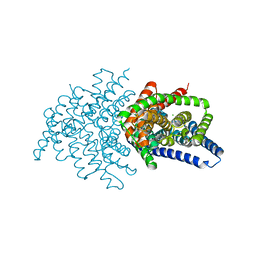 | | Crystal structure of ClC-ec1 triple mutant (E113Q, E148Q, E203Q) | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Maduke, M, Mathews, I.I, Chavan, T.S. | | Deposit date: | 2019-11-24 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl - /H + transport cycle.
Elife, 9, 2020
|
|
