6FFI
 
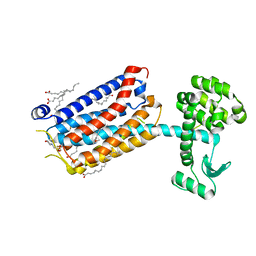 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
8OJR
 
 | | Solution NMR Structure of Alpha-Synuclein 1-25 Peptide in 50% TFE. | | Descriptor: | Alpha-synuclein | | Authors: | Allen, S.G, Williams, C, Meade, R.M, Tang, T.M.S, Crump, M.P, Mason, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | An N-terminal alpha-Synuclein fragment binds lipid vesicles to modulate lipid induced aggregation
Cell Rep Phys Sci, 2023
|
|
8OL8
 
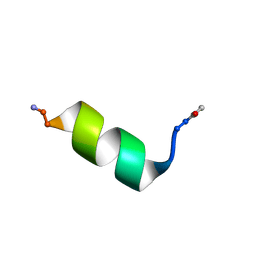 | |
2Y44
 
 | | Crystal structure of GARP from Trypanosoma congolense | | Descriptor: | GLUTAMIC ACID/ALANINE-RICH PROTEIN, GLYCEROL, IODIDE ION | | Authors: | Loveless, B.C, Mason, J.W, Sakurai, T, Inoue, N, Razavi, M, Pearson, T.W, Boulanger, M.J. | | Deposit date: | 2011-01-04 | | Release date: | 2011-03-30 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization and Epitope Mapping of the Glutamic Acid/Alanine-Rich Protein from Trypanosoma Congolense: Defining Assembly on the Parasite Cell Surface.
J.Biol.Chem., 286, 2011
|
|
5LWE
 
 | | Crystal structure of the human CC chemokine receptor type 9 (CCR9) in complex with vercirnon | | Descriptor: | C-C chemokine receptor type 9, CHOLESTEROL, MALONATE ION, ... | | Authors: | Oswald, C, Rappas, M, Kean, J, Dore, A.S, Errey, J.C, Bennett, K, Deflorian, F, Christopher, J.A, Jazayeri, A, Mason, J.S, Congreve, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intracellular allosteric antagonism of the CCR9 receptor.
Nature, 540, 2016
|
|
6ZDR
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 4d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6ZDV
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 5d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5FV8
 
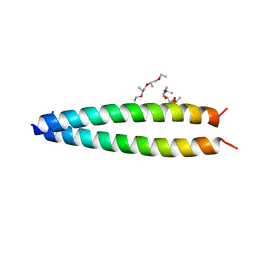 | |
6GIK
 
 | | NMR structure of temporin B L1FK in SDS micelles | | Descriptor: | temporinB_L1FK | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GS5
 
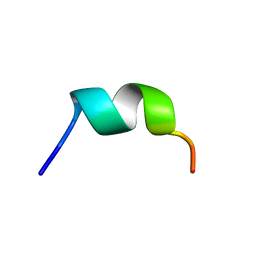 | | NMR structure of temporin L in SDS micelles | | Descriptor: | Temporin-L | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | Descriptor: | Temporin-B | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GS9
 
 | | NMR structure of aurein 2.5 in SDS micelles | | Descriptor: | Aurein 2.5 | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GIJ
 
 | | NMR structure of temporin B KKG6A in SDS micelles | | Descriptor: | temporinB_KKG6A | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6FFH
 
 | | Crystal Structure of mGluR5 in complex with Fenobam at 2.65 A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(3-chlorophenyl)-3-(3-methyl-5-oxidanylidene-4~{H}-imidazol-2-yl)urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
7RQ8
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
7RQ9
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
5NX2
 
 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | Authors: | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-05-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
4AQC
 
 | | Triazolopyridine-based Inhibitor of Janus Kinase 2 | | Descriptor: | 8-(4-methylsulfonylphenyl)-N-(4-morpholin-4-ylphenyl)-[1,2,4]triazolo[1,5-a]pyridin-2-amine, SULFATE ION, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Dugan, B.J, Gingrich, D.E, Mesaros, E.F, Milkiewicz, K.L, Curry, M.A, Zulli, A.L, Dobrzanski, P, Serdikoff, C, Jan, M, Angeles, T.S, Albom, M.S, Mason, J.L, Aimone, L.D, Meyer, S.L, Huang, Z, Wells-Knecht, K.J, Ator, M.A, Ruggeri, B.A, Dorsey, B.D. | | Deposit date: | 2012-04-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Selective, Orally Bioavailable 1,2,4-Triazolo[1,5-A]Pyridine-Based Inhibitor of Janus Kinase 2 for Use in Anticancer Therapy: Discovery of Cep-33779.
J.Med.Chem., 55, 2012
|
|
3UZA
 
 | | Thermostabilised Adenosine A2A receptor in complex with 6-(2,6-Dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine | | Descriptor: | 6-(2,6-dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine, Adenosine receptor A2a | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Tehan, B, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.273 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
3UZC
 
 | | Thermostabilised Adenosine A2A receptor in complex with 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol | | Descriptor: | 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol, Adenosine A2A Receptor | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.341 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
2LS9
 
 | | Pleurocidin-NH2 | | Descriptor: | Pleurocidin | | Authors: | Vermeer, L.S, Kozlowska, J, Mason, J.A. | | Deposit date: | 2012-04-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | All Atom Simulations of the Initial Binding of Magainin and Pleurocidin to Membranes Comprising of a Mixture of Anionic and Zwitterionic Lipids
To be Published
|
|
2L99
 
 | | Solution structure of LAK160-P10 | | Descriptor: | LAK160-P10 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L9A
 
 | | Solution structure of LAK160-P12 | | Descriptor: | LAK160-P12 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2LSA
 
 | | Magainin | | Descriptor: | Magainin-2 | | Authors: | Vermeer, L.S, Mason, J.A. | | Deposit date: | 2012-04-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | All Atom Simulations of the Initial Binding of Magainin and Pleurocidin to Membranes Comprising a Mixture of Anionic and Zwitterionic Lipids
To be Published
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
