5DAD
 
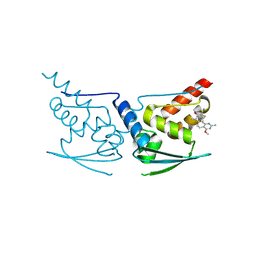 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64014 | | Descriptor: | (6aS,7S,10aS)-8-hydroxy-4-methoxy-2,7,10a-trimethyl-5,6,6a,7,10,10a-hexahydrobenzo[h]quinazoline-9-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
5FKJ
 
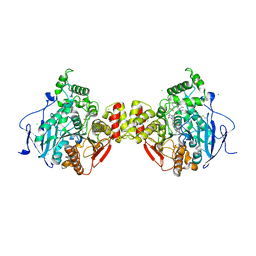 | | Crystal structure of mouse acetylcholinesterase in complex with C-547, an alkyl ammonium derivative of 6-methyl uracil | | Descriptor: | 1,3-BIS[5(DIETHYL-O-NITROBENZYLAMMONIUM)PENTYL]-6-METHYLURACIL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Nachon, F, Villard-Wandhammer, M, Petrov, K, Masson, P. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Slow-Binding Inhibition of Acetylcholinesterase by a 6-Methyluracil Alkyl-Ammonium Derivative: Mechanism and Advantages for Myasthenia Gravis Treatment.
Biochem.J., 473, 2016
|
|
5FYL
 
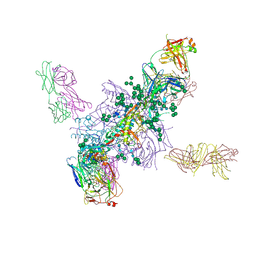 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5FYJ
 
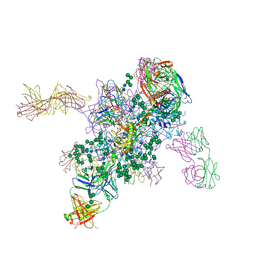 | | Crystal Structure at 3.4 A Resolution of Fully Glycosylated HIV-1 Clade G X1193.c1 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5FYK
 
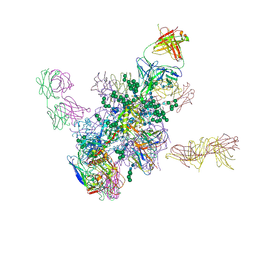 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade B JR-FL SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
3ISN
 
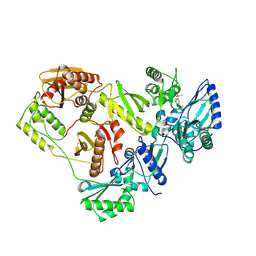 | | Crystal structure of HIV-1 RT bound to A 6-vinylpyrimidine inhibitor | | Descriptor: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ennifar, E, Freisz, S, Bec, G, Dumas, P, Botta, M, Radi, M. | | Deposit date: | 2009-08-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3WI5
 
 | | Crystal structure of the Loop 7 mutant PorB from Neisseria meningitidis serogroup B | | Descriptor: | CITRATE ANION, Major outer membrane protein P.IB | | Authors: | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
3ITH
 
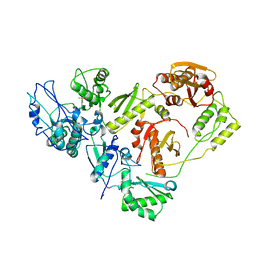 | | Crystal structure of the HIV-1 reverse transcriptase bound to a 6-vinylpyrimidine inhibitor | | Descriptor: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Freisz, S, Bec, G, Wolff, P, Dumas, P, Radi, M, Botta, M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3SI7
 
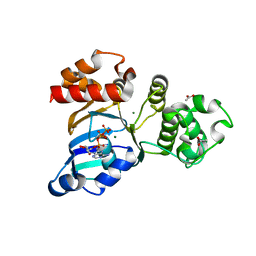 | | The crystal structure of the NBD1 domain of the mouse CFTR protein, deltaF508 mutant | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Brautigam, C.A, Caspa, E, Thomas, P.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Requirements for efficient correction of DeltaF508 CFTR revealed by analyses of evolved sequences
Cell(Cambridge,Mass.), 148, 2012
|
|
462D
 
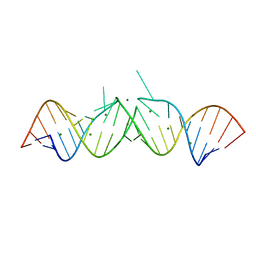 | | CRYSTAL STRUCTURE OF THE HIV-1 GENOMIC RNA DIMERIZATION INITIATION SITE | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G) -3') | | Authors: | Ennifar, E, Yusupov, M, Walter, P, Marquet, R, Ehresmann, C, Ehresmann, B, Dumas, P. | | Deposit date: | 1999-03-18 | | Release date: | 1999-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges.
Structure Fold.Des., 7, 1999
|
|
4TRA
 
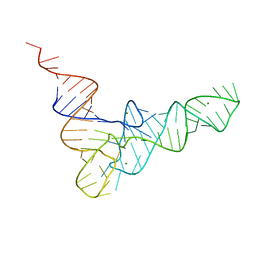 | |
3Q4L
 
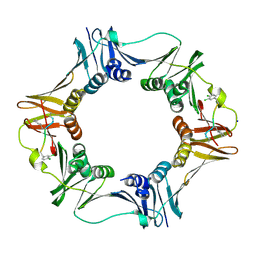 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3WI4
 
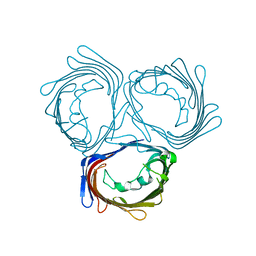 | | Crystal structure of wild-type PorB from Neisseria meningitidis serogroup B | | Descriptor: | Major outer membrane protein P.IB | | Authors: | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
3TRA
 
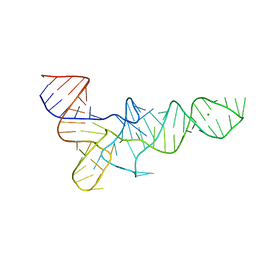 | |
4BC1
 
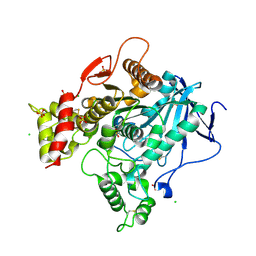 | | Structure of mouse acetylcholinesterase inhibited by CBDP (30-min soak): cresyl-saligenin-phosphoserine adduct | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
3Q4J
 
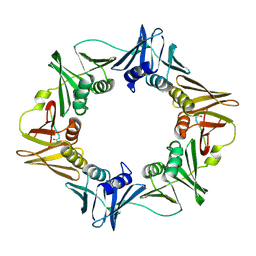 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
4BBZ
 
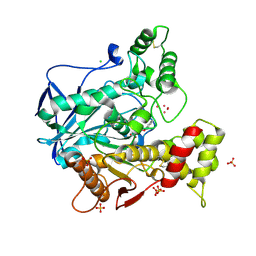 | | Structure of human butyrylcholinesterase inhibited by CBDP (2-min soak): Cresyl-phosphoserine adduct | | Descriptor: | (2-methylphenyl) dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
4BC0
 
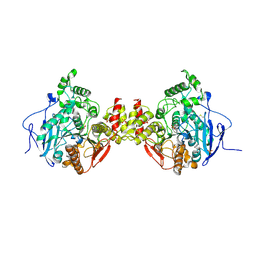 | | Structure of mouse acetylcholinesterase inhibited by CBDP (12-h soak) : Cresyl-phosphoserine adduct | | Descriptor: | (2-methylphenyl) dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
3Q4K
 
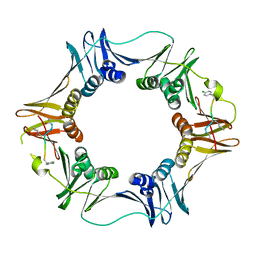 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3DHB
 
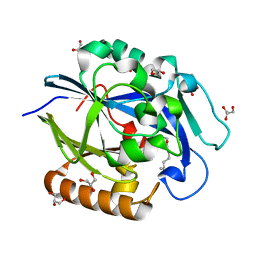 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DL7
 
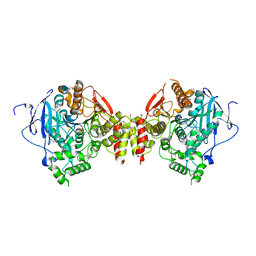 | | Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
3D65
 
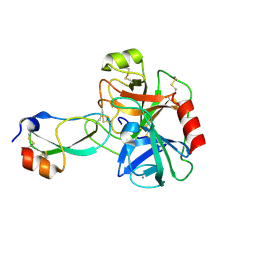 | | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Textilinin | | Authors: | Millers, E.-K.I, Masci, P.P, Lavin, M.F, de Jersey, J, Guddat, L.W. | | Deposit date: | 2008-05-19 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin
To be Published
|
|
3DW4
 
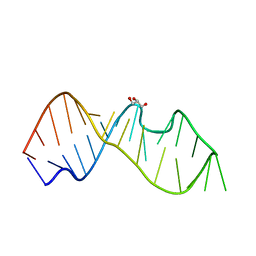 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DVZ
 
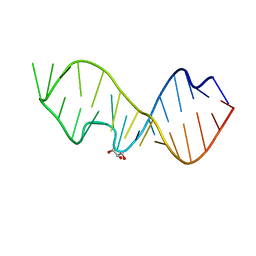 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
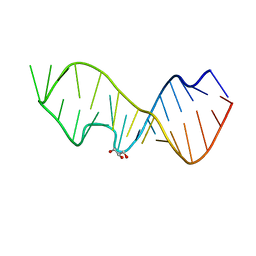 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
