1T2C
 
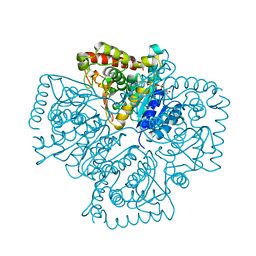 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1J1A
 
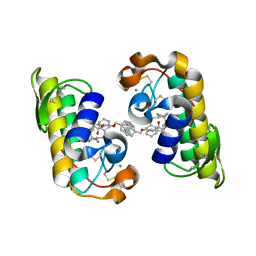 | | PANCREATIC SECRETORY PHOSPHOLIPASE A2 (IIa) WITH ANTI-INFLAMMATORY ACTIVITY | | Descriptor: | (S)-5-(4-BENZYLOXY-PHENYL)-4-(7-PHENYL-HEPTANOYLAMINO)-PENTANOIC ACID, CALCIUM ION, Phospholipase A2 | | Authors: | Hansford, K.A, Reid, R.C, Clark, C.I, Tyndall, J.D.A, Whitehouse, M.W, Guthrie, T, McGeary, R.P, Schafer, K, Martin, J.L, Fairlie, D.P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D-Tyrosine as a Chiral Precusor to Potent Inhibitors of Human Nonpancreatic Secretory Phospholipase A2 (IIa) with Antiinflammatory Activity
Chembiochem, 4, 2003
|
|
1T25
 
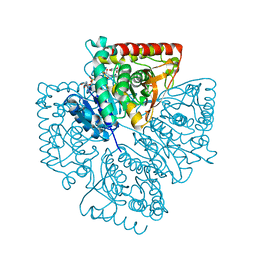 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH and 3-hydroxyisoxazole-4-carboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-HYDROXYISOXAZOLE-4-CARBOXYLIC ACID, GLYCEROL, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
2V1O
 
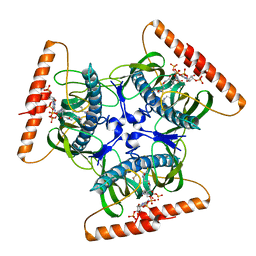 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1T2E
 
 | | Plasmodium falciparum lactate dehydrogenase S245A, A327P mutant complexed with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1T2D
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with NAD+ and oxalate | | Descriptor: | GLYCEROL, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1T2F
 
 | | Human B lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | | Descriptor: | 4-HYDROXY-1,2,5-OXADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase B chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1LS6
 
 | | Human SULT1A1 complexed with PAP and p-Nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, aryl sulfotransferase | | Authors: | Gamage, N.U, Barnett, A.C, Tresillian, M, Latham, C.F, Liyou, N.E, McManus, M.E, Martin, J.L. | | Deposit date: | 2002-05-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition.
J.Biol.Chem., 278, 2003
|
|
1T26
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH and 4-hydroxy-1,2,5-thiadiazole-3-carboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-HYDROXY-1,2,5-THIADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
3PUJ
 
 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2WFW
 
 | | Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases - The Arc domain structure | | Descriptor: | ARC | | Authors: | Djuranovic, S, Hartmann, M.D, Habeck, M, Ursinus, A, Zwickl, P, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
4MLY
 
 | | Disulfide isomerase from multidrug resistance IncA/C related integrative and conjugative elements in oxidized state (P21 space group) | | Descriptor: | 1,3-BUTANEDIOL, DsbP | | Authors: | Premkumar, L, Kurth, F, Neyer, S, Martin, J.L. | | Deposit date: | 2013-09-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | The Multidrug Resistance IncA/C Transferable Plasmid Encodes a Novel Domain-swapped Dimeric Protein-disulfide Isomerase.
J.Biol.Chem., 289, 2014
|
|
2VBU
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBV
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and FMN | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBT
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and PO4 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBS
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with PO4 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Djuranovic, S, Ammelburg, M, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
1UTE
 
 | | PIG PURPLE ACID PHOSPHATASE COMPLEXED WITH PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, MU-OXO-DIIRON, ... | | Authors: | Guddat, L.W, Mcalpine, A, Hume, D, Hamilton, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1999-01-18 | | Release date: | 1999-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mammalian purple acid phosphatase.
Structure Fold.Des., 7, 1999
|
|
4MCU
 
 | |
4ML6
 
 | |
4ML1
 
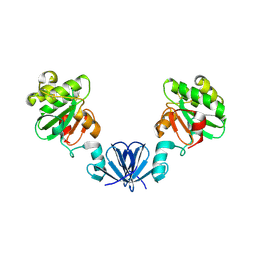 | |
2H0G
 
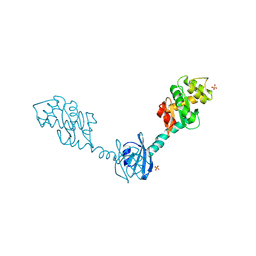 | | Crystal Structure of DsbG T200M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
2H0H
 
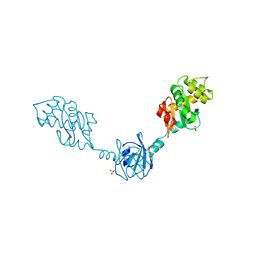 | | Crystal Structure of DsbG K113E mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
2H0I
 
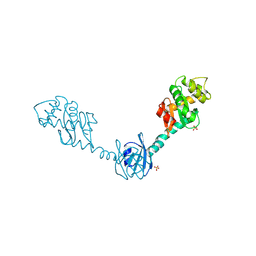 | | Crystal Structure of DsbG V216M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
4K6X
 
 | | Crystal structure of disulfide oxidoreductase from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disulfide oxidoreductase | | Authors: | Premkumar, L, Martin, J.L. | | Deposit date: | 2013-04-16 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6TLZ
 
 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 3'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
