1PVP
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVQ
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | DNA 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVR
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | Descriptor: | 34-MER, Recombinase CRE | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
6BHN
 
 | | Red Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Yu, Q, Lim, S, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5AJM
 
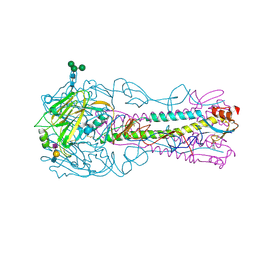 | | H5 (VN1194) Asn186Lys Mutant Haemagglutinin in Complex with Avian Receptor Analogue 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
6BHO
 
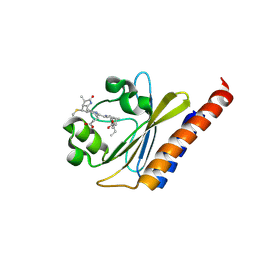 | | Green Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Lim, S, Yu, Q, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1KWM
 
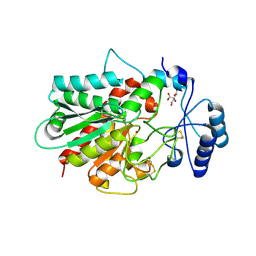 | | Human procarboxypeptidase B: Three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI) | | Descriptor: | CITRIC ACID, Procarboxypeptidase B, ZINC ION | | Authors: | Pereira, P.J.B, Segura-Martin, S, Ferrer-Orta, C, Vendrell, J, Aviles, F.-X, Coll, M, Gomis-Rueth, F.-X. | | Deposit date: | 2002-01-30 | | Release date: | 2002-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human procarboxypeptidase B: three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI).
J.Mol.Biol., 321, 2002
|
|
5JW4
 
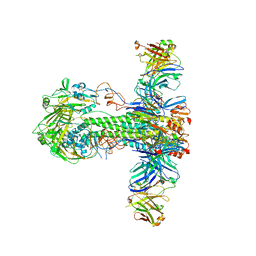 | | Structure of MEDI8852 Fab Fragment in Complex with H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Neu, U, Collins, P.J, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
5JW5
 
 | | Structure of MEDI8852 Fab Fragment | | Descriptor: | MEDI8852 Heavy chain, MEDI8852 Light chain, PHOSPHATE ION | | Authors: | Neu, U, Collins, P.J, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
5JW3
 
 | | Structure of MEDI8852 Fab Fragment in Complex with H7 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Collins, P.J, Neu, U, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
2BQZ
 
 | | Crystal structure of a ternary complex of the human histone methyltransferase Pr-SET7 (also known as SET8) | | Descriptor: | HISTONE H4, S-ADENOSYL-L-HOMOCYSTEINE, SET8 PROTEIN | | Authors: | Xiao, B, Jing, C, Kelly, G, Walker, P.A, Muskett, F.W, Frenkiel, T.A, Martin, S.R, Sarma, K, Reinberg, D, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Genes Dev., 19, 2005
|
|
4Z38
 
 | |
4Q0N
 
 | | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(2,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridin-6-yl)prop-2-en-1-one, 1,2-ETHANEDIOL, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Monteiro, O, Fedorov, O, Chaikuad, A, Yue, W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand
To be Published
|
|
3IJC
 
 | | Crystal structure of Eed in complex with NDSB-195 | | Descriptor: | ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
2BF0
 
 | | crystal structure of the rpr of pcf11 | | Descriptor: | CALCIUM ION, PCF11 | | Authors: | Noble, C.G, Hollingworth, D, Martin, S.R, Adeniran, V.E, Smerdon, S.J, Kelly, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2004-12-02 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Key Features of the Interaction between Pcf11 Cid and RNA Polymerase II Ctd.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7ONS
 
 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
2ATW
 
 | | Structure of a Mycobacterium tuberculosis NusA-RNA complex | | Descriptor: | Transcription elongation protein nusA, ribosomal RNA (5'- AGAACUCAAUAG -3') | | Authors: | Beuth, B, Pennell, S, Arnvig, K.B, Martin, S.R, Taylor, I.A. | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a Mycobacterium tuberculosis NusA-RNA complex.
Embo J., 24, 2005
|
|
4UNW
 
 | | Structure of the A_Equine_Newmarket_2_93 H3 haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Haire, L.F, Ogrodowicz, R.W, Martin, S.R, Walker, P.A, Xiong, X, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recent Evolution of Equine Influenza and the Origin of Canine Influenza.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4UO7
 
 | | Structure of the A_Canine_Colorado_17864_06 H3 haemagglutinin in complex with 6SO4 Sialyl Lewis X | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ HA1, ... | | Authors: | Vachieri, S.G, Collins, P.J, Haire, L.F, Ogrodowicz, R.W, Martin, S.R, Walker, P.A, Xiong, X, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recent Evolution of Equine Influenza and the Origin of Canine Influenza.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3IIY
 
 | | Crystal structure of Eed in complex with a trimethylated histone H1K26 peptide | | Descriptor: | Histone H1K26 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IIW
 
 | | Crystal structure of Eed in complex with a trimethylated histone H3K27 peptide | | Descriptor: | Histone H3 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ0
 
 | | Crystal structure of Eed in complex with a trimethylated histone H3K9 peptide | | Descriptor: | Histone H3K9 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ1
 
 | | Crystal structure of Eed in complex with a trimethylated histone H4K20 peptide | | Descriptor: | Histone H4K20 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
