1HU8
 
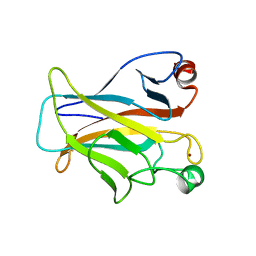 | | CRYSTAL STRUCTURE OF THE MOUSE P53 CORE DNA-BINDING DOMAIN AT 2.7A RESOLUTION | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Zhao, K, Chai, X, Johnston, K, Clements, A, Marmorstein, R. | | Deposit date: | 2001-01-04 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution.
J.Biol.Chem., 276, 2001
|
|
6O07
 
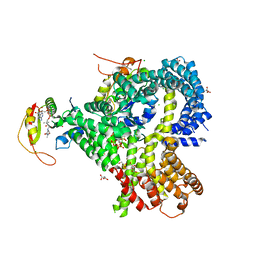 | |
3BY4
 
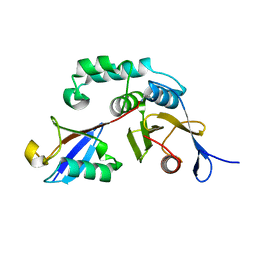 | |
3C0R
 
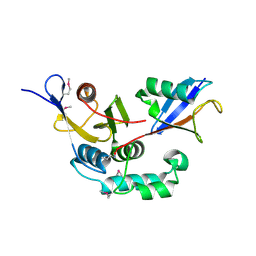 | |
3LBX
 
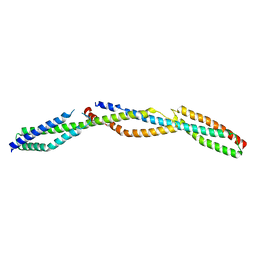 | | Crystal Structure of the Erythrocyte Spectrin Tetramerization Domain Complex | | Descriptor: | Spectrin alpha chain, erythrocyte, Spectrin beta chain | | Authors: | Ipsaro, J.J, Harper, S.L, Messick, T.E, Marmorstein, R, Mondragon, A, Speicher, D.W. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex.
Blood, 115, 2010
|
|
4DAW
 
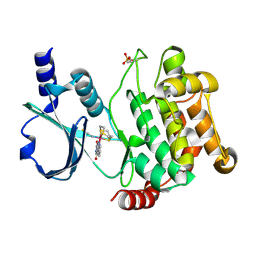 | | Crystal structure of PAK1 kinase domain with the ruthenium phthalimide complex | | Descriptor: | Serine/threonine-protein kinase PAK 1, [1,3-dioxo-6-(pyridin-2-yl-kappaN)-2,3-dihydro-1H-isoindol-5-yl-kappaC~5~][(thioxomethylidene)azanido-kappaN](1,4,7-trithionane-kappa~3~S~1~,S~4~,S~7~)ruthenium | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The art of filling protein pockets efficiently with octahedral metal complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8ETB
 
 | | the crystal structure of a rationally designed zinc sensor based on maltose binding protein - Zn binding conformation | | Descriptor: | ACETATE ION, ZINC ION, Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.D, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
8F23
 
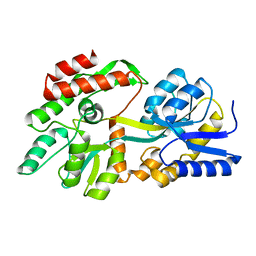 | | The crystal structure of a rationally designed zinc sensor based on maltose binding protein - Apo conformation | | Descriptor: | Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.d, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
3FXZ
 
 | |
5WJD
 
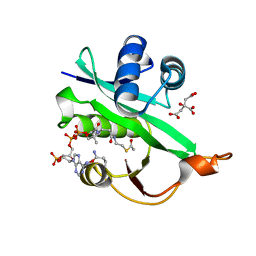 | | Crystal structure of Naa80 bound to acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, CG8481, isoform B, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
9B3T
 
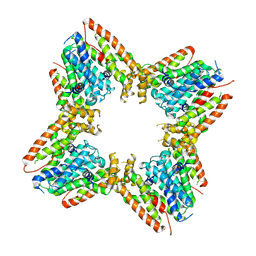 | |
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TRO
 
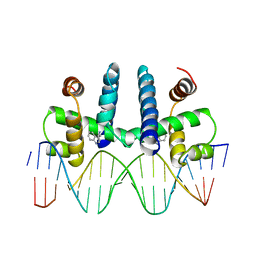 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
1SZC
 
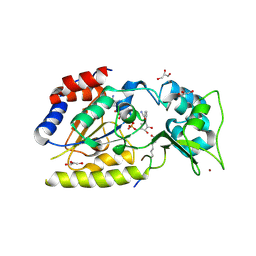 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1IHB
 
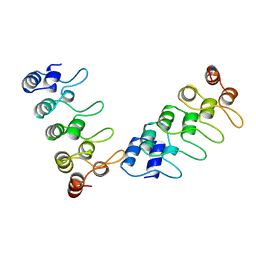 | | CRYSTAL STRUCTURE OF P18-INK4C(INK6) | | Descriptor: | CYCLIN-DEPENDENT KINASE 6 INHIBITOR | | Authors: | Ravichandran, V, Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-10-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations.
Nat.Struct.Biol., 5, 1998
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
5ICV
 
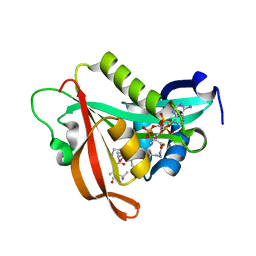 | | Crystal structure of human NatF (hNaa60) bound to a bisubstrate analogue | | Descriptor: | MET-LYS-ALA-VAL-LIG, N-alpha-acetyltransferase 60, [5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)furan-2-yl]methyl (3R)-4-{[3-({(E)-2-[(2,2-dihydroxyethyl)sulfanyl]ethenyl}amino)-3-oxopropyl]amino}-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
5ICW
 
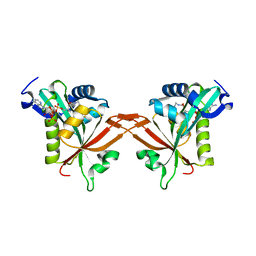 | | Crystal structure of human NatF (hNaa60) homodimer bound to Coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, N-alpha-acetyltransferase 60 | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
6POE
 
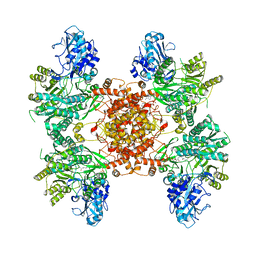 | | Structure of ACLY in complex with CoA | | Descriptor: | ATP-citrate synthase, COENZYME A | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1K6O
 
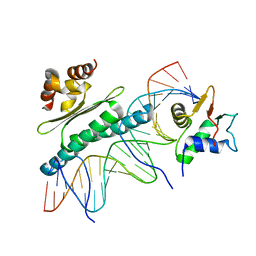 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
8G0L
 
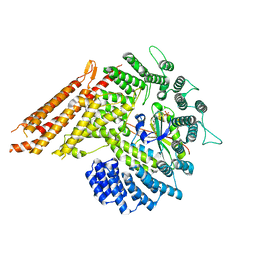 | |
3Q35
 
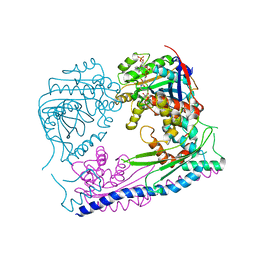 | | Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, Histone acetyltransferase, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
3Q33
 
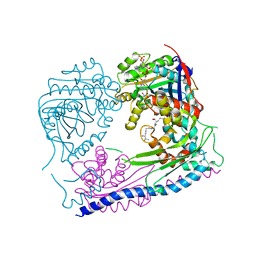 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
1ZX2
 
 | | Crystal Structure of Yeast UBP3-associated Protein BRE5 | | Descriptor: | UBP3-associated protein BRE5 | | Authors: | Li, K, Zhao, K, Ossareh-Nazari, B, Da, G, Dargemont, C, Marmorstein, R. | | Deposit date: | 2005-06-06 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor
J.Biol.Chem., 280, 2005
|
|
2QIY
 
 | |
