3TO6
 
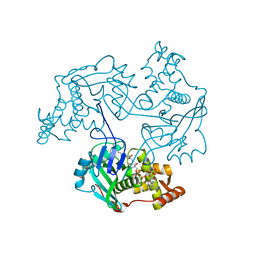 | |
3QM0
 
 | | Crystal structure of RTT109-AC-CoA complex | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | Authors: | Tang, Y, Marmorstein, R. | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-16 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
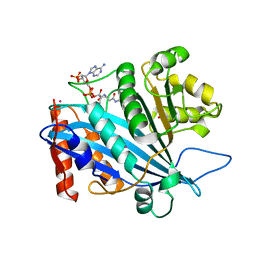
3TO9
 
 | | Crystal structure of yeast Esa1 E338Q HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | 1,2-ETHANEDIOL, CACODYLIC ACID, COENZYME A, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
4R5L
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-C) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9701 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5J
 
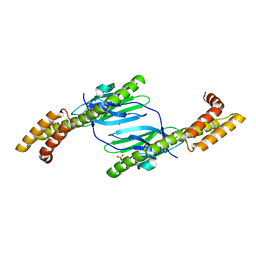 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-A) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5I
 
 | | Crystal structure of the DnaK C-terminus with the substrate peptide NRLLLTG | | Descriptor: | Chaperone protein DnaK, HSP70/DnaK Substrate Peptide: NRLLLTG, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9702 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5G
 
 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
7L1K
 
 | |
3TFY
 
 | |
2FQ3
 
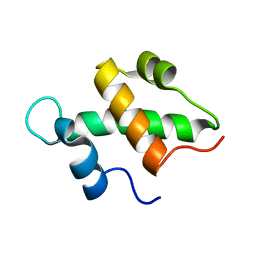 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | Descriptor: | Transcription regulatory protein SWI3 | | Authors: | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3FY0
 
 | |
2HAP
 
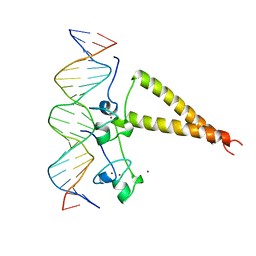 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
2IOO
 
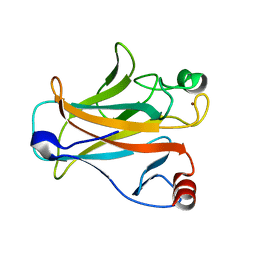 | | Crystal structure of the mouse p53 core domain | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOI
 
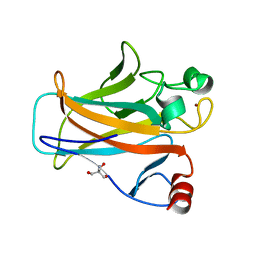 | | Crystal structure of the mouse p53 core domain at 1.55 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOM
 
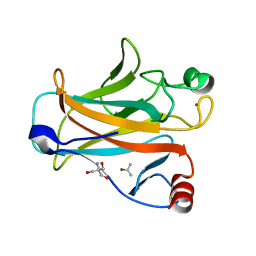 | | Mouse p53 core domain soaked with 2-propanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ISOPROPYL ALCOHOL, ... | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3CO7
 
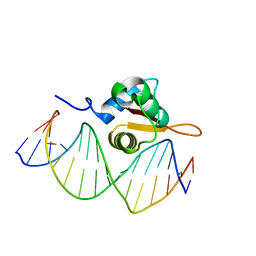 | | Crystal Structure of FoxO1 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DAP*DAP*DAP*DTP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DGP*DA)-3'), DNA (5'-D(*DTP*DCP*DTP*DTP*DGP*DTP*DTP*DTP*DAP*DCP*DAP*DTP*DTP*DTP*DTP*DG)-3'), Forkhead box protein O1 | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
2GEQ
 
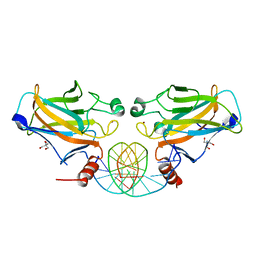 | | Crystal Structure of a p53 Core Dimer Bound to DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*GP*TP*GP*AP*GP*CP*AP*TP*GP*CP*TP*CP*AP*C)-3', Cellular tumor antigen p53, ... | | Authors: | Ho, W.C, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-03-20 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the p53 Core Domain Dimer Bound to DNA.
J.Biol.Chem., 281, 2006
|
|
3COA
 
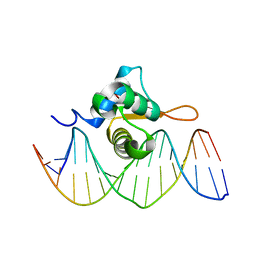 | | Crystal Structure of FoxO1 DBD Bound to IRE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DCP*DAP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DGP*DTP*DTP*DTP*DGP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DG)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3CO6
 
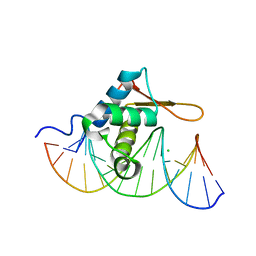 | | Crystal Structure of FoxO1 DBD Bound to DBE1 DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3DM7
 
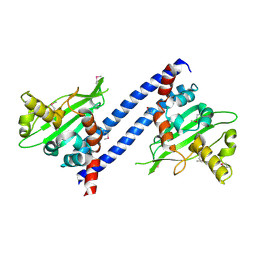 | |
3CSF
 
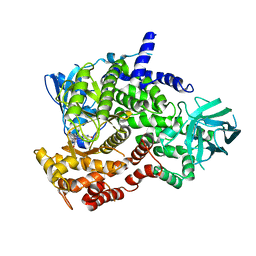 | |
3CST
 
 | |
1Q1A
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, HST2 protein, Histone H4, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-Acetyl ADP ribose and histone peptide.
Structure, 11, 2003
|
|
1Q17
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, HST2 protein, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Yeast Hst2 Protein Deacetylase in Ternary Complex with 2'-O-Acetyl
ADP Ribose and Histone Peptide.
Structure, 11, 2003
|
|
1O9K
 
 | | Crystal structure of the retinoblastoma tumour suppressor protein bound to E2F peptide | | Descriptor: | RETINOBLASTOMA-ASSOCIATED PROTEIN, TRANSCRIPTION FACTOR E2F1 | | Authors: | Xiao, B, Spencer, J, Clements, A, Ali-Khan, N, Mittnacht, S, Broceno, C, Burghammer, M, Perrakis, A, Marmorstein, R, Gamblin, S.J. | | Deposit date: | 2002-12-16 | | Release date: | 2003-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Retinoblastoma Tumor Suppressor Protein Bound to E2F and the Molecular Basis of its Regulation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
