5D2N
 
 | | Crystal structure of C25-NLV-HLA-A2 complex | | Descriptor: | ASN-LEU-VAL-PRO-MET-VAL-ALA-THR-VAL, Beta-2-microglobulin, C25 alpha, ... | | Authors: | Mariuzza, R.A, Yang, X. | | Deposit date: | 2015-08-05 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for Clonal Diversity of the Public T Cell Response to a Dominant Human Cytomegalovirus Epitope.
J.Biol.Chem., 290, 2015
|
|
1NBZ
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K97A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NDM
 
 | |
1NBY
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K96A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
1NDG
 
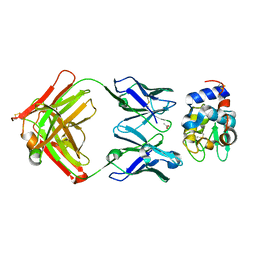 | | Crystal structure of Fab fragment of antibody HyHEL-8 complexed with its antigen lysozyme | | Descriptor: | ACETIC ACID, Lysozyme C, antibody kappa light chain, ... | | Authors: | Mariuzza, R.A, Li, Y, Li, H, Yang, F, Smith-Gill, S.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of the maturation of an antibody response to a protein antigen
Nat.Struct.Biol., 10, 2003
|
|
3T0E
 
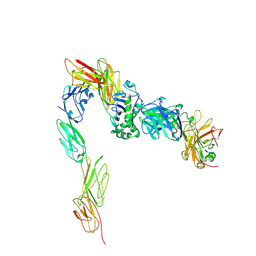 | | Crystal structure of a complete ternary complex of T cell receptor, peptide-MHC and CD4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Yin, Y, Mariuzza, R.A. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4E41
 
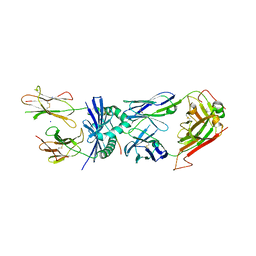 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
4E42
 
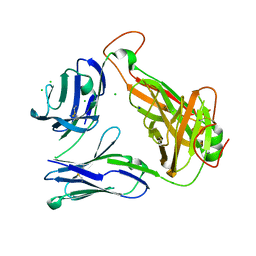 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | CHLORIDE ION, NITRATE ION, SODIUM ION, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
3FF8
 
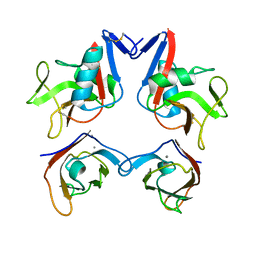 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | CALCIUM ION, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
8FWH
 
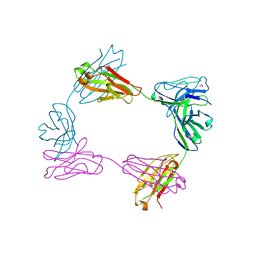 | | Crystal structure of bivalent antibody Fab fragment of Anti-human LAG3 (22D2) | | Descriptor: | 1,2-ETHANEDIOL, Anti-human LAG3 (22D2) heavy chain, Anti-human LAG3 (22D2) light chain | | Authors: | Mishra, A.K, Agnihotri, P, Mariuzza, R.A. | | Deposit date: | 2023-01-22 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
4Z0X
 
 | |
4ZSO
 
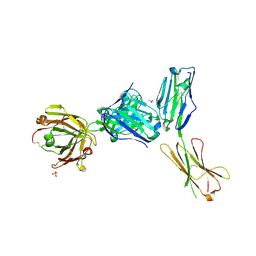 | | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, X, Li, Y, Mariuzza, R.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody
to be published
|
|
8GOP
 
 | | SARS-CoV-2 specific private TCR RLQ7 | | Descriptor: | SARS-CoV-2 specific private TCR RLQ7 alpha, SARS-CoV-2 specific private TCR RLQ7 beta | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
8GOM
 
 | | SARS-CoV-2 specific private TCR RLQ7 in complex with RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 specific private TCR RLQ7 alpha, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
8GON
 
 | |
4XVJ
 
 | |
8SR0
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8SO3
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8YJ3
 
 | | N17.1.2 recognition of NRAS neoantigens | | Descriptor: | tcr alpha, tcr beta | | Authors: | Wu, D.C, Mariuzza, R.A. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Sci Adv, 10, 2024
|
|
8YJ2
 
 | | N17.1.2 recognition of NRAS neoantigens | | Descriptor: | Beta-2-microglobulin, ILE-LEU-ASP-THR-ALA-GLY-ARG-GLU-GLU-TYR, human leukocyte antigen, ... | | Authors: | Wu, D.C, Mariuzza, R.A. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Sci Adv, 10, 2024
|
|
8YIV
 
 | | N17.1.2 recognition of NRAS neoantigens | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Wu, D.C, Mariuzza, R.A. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Sci Adv, 10, 2024
|
|
1YWP
 
 | | Phospholipase Cgamma1 SH3 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 | | Authors: | Deng, L, Velikovsky, C.A, Swaminathan, C.P, Cho, S, Mariuzza, R.A. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Recognition of the T Cell Adaptor Protein SLP-76 by the SH3 Domain of Phospholipase Cgamma1
J.Mol.Biol., 352, 2005
|
|
1ZGL
 
 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
7R6J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 1 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[3-methyl-5-(pyrimidin-2-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7REV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
