8C83
 
 | | Cryo-EM structure of in vitro reconstituted Otu2-bound Ub-40S complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11-A, ... | | 著者 | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2023-01-18 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
4PHK
 
 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids. The complex with (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid | | 分子名称: | (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid, CALCIUM ION, Calpain small subunit 1 | | 著者 | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B, Robinson, E. | | 登録日 | 2014-05-06 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2018-12-16 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8CBJ
 
 | | Cryo-EM structure of Otu2-bound cytoplasmic pre-40S ribosome biogenesis complex | | 分子名称: | 20S pre-ribosomal RNA, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | 著者 | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2023-01-25 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAS
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2023-01-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Dybkov, O, Kastner, B, Luehrmann, R. | | 登録日 | 2023-01-12 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
5NV9
 
 | | Substrate-bound outward-open state of a Na+-coupled sialic acid symporter reveals a novel Na+-site | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, N-acetyl-beta-neuraminic acid, PHOSPHATE ION, ... | | 著者 | Wahlgren, W.Y, North, R.A, Dunevall, E, Paz, A, Goyal, P, Bisignano, P, Grabe, M, Dobson, R, Abramson, J, Ramaswamy, S, Friemann, R. | | 登録日 | 2017-05-03 | | 公開日 | 2018-04-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+site.
Nat Commun, 9, 2018
|
|
6H4E
 
 | | Proteus mirabilis N-acetylneuraminate lyase | | 分子名称: | Putative N-acetylneuraminate lyase, SULFATE ION | | 著者 | North, R.A, Garcia-Bonete, M.J, Goyal, P, Katona, G, Dobson, R.C.J, Friemann, R. | | 登録日 | 2018-07-21 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.561 Å) | | 主引用文献 | The structure of Proteus mirabilis N-acetylneuraminate lyase reveals an intermolecular disulphide bond
To Be Published
|
|
3G4D
 
 | | Crystal Structure of (+)-delta-Cadinene Synthase from Gossypium arboreum and Evolutionary Divergence of Metal Binding Motifs for Catalysis | | 分子名称: | (+)-delta-cadinene synthase isozyme XC1, BETA-MERCAPTOETHANOL, GLYCEROL | | 著者 | Gennadios, H.A, Di Costanzo, L, Miller, D.J, Allemann, R.K, Christianson, D.W. | | 登録日 | 2009-02-03 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
8BQD
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BQX
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
3IZQ
 
 | | Structure of the Dom34-Hbs1-GDPNP complex bound to a translating ribosome | | 分子名称: | Elongation factor 1 alpha-like protein, Protein DOM34 | | 著者 | Becker, T, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Sieber, H, Abdel Motaal, B, Mielke, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2010-11-30 | | 公開日 | 2011-06-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | 著者 | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | 登録日 | 2014-04-17 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
5NWY
 
 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | 登録日 | 2017-05-08 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
6FTJ
 
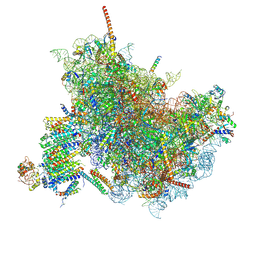 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome | | 分子名称: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Braunger, K, Becker, T, Beckmann, R. | | 登録日 | 2018-02-22 | | 公開日 | 2018-03-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
3J2I
 
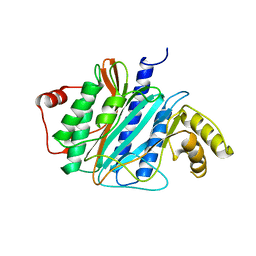 | | Structure of late pre-60S ribosomal subunits with nuclear export factor Arx1 bound at the peptide exit tunnel | | 分子名称: | Eukaryotic translation initiation factor 6, Proliferation-associated protein 2G4 | | 著者 | Bradatsch, B, Leidig, C, Granneman, S, Gnaedig, M, Tollervey, D, Boettcher, B, Beckmann, R, Hurt, E. | | 登録日 | 2012-10-04 | | 公開日 | 2012-11-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.9 Å) | | 主引用文献 | Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4Q5Y
 
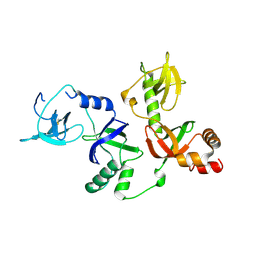 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | 分子名称: | Maternal protein tudor | | 著者 | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | 登録日 | 2014-04-18 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
1MZP
 
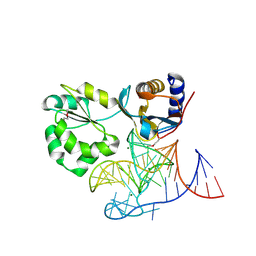 | | Structure of the L1 protuberance in the ribosome | | 分子名称: | 50s ribosomal protein L1P, MAGNESIUM ION, fragment of 23S rRNA | | 著者 | Nikulin, A, Eliseikina, I, Tishchenko, S, Nevskaya, N, Davydova, N, Platonova, O, Piendl, W, Selmer, M, Liljas, A, Zimmermann, R, Garber, M, Nikonov, S. | | 登録日 | 2002-10-09 | | 公開日 | 2003-01-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of the L1 protuberance in the ribosome.
Nat.Struct.Biol., 10, 2003
|
|
1NPA
 
 | | crystal structure of HIV-1 protease-hup | | 分子名称: | (3S)-TETRAHYDROFURAN-3-YL (1R,2S)-3-[4-((1R)-2-{[(S)-AMINO(HYDROXY)METHYL]OXY}-2,3-DIHYDRO-1H-INDEN-1-YL)-2-BENZYL-3-OXOPYRROLIDIN-2-YL]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, POL polyprotein | | 著者 | Smith III, A.B, Hirschmann, R, Pasternak, A, Yao, W, Sprengeler, P.A, Holloway, M.K, Kuo, L.C, Chen, Z, Darke, P.L, Schleif, W.A. | | 登録日 | 2003-01-17 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An orally bioavailable pyrrolinone inhibitor of HIV-1 protease: computational analysis and X-ray crystal structure of the enzyme complex.
J.MED.CHEM., 40, 1997
|
|
3J25
 
 | | Structural basis for TetM-mediated tetracycline resistance | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tetracycline resistance protein tetM | | 著者 | Doenhoefer, A, Franckenberg, S, Wickles, S, Berninghausen, O, Beckmann, R, Wilson, D.N. | | 登録日 | 2012-08-22 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Structural basis for TetM-mediated tetracycline resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1V1D
 
 | |
6EM5
 
 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | 登録日 | 2017-10-01 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
1E7K
 
 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | 分子名称: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | 著者 | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | 登録日 | 2000-08-29 | | 公開日 | 2001-02-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
6FF7
 
 | | human Bact spliceosome core structure | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | 著者 | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | 登録日 | 2018-01-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
8R55
 
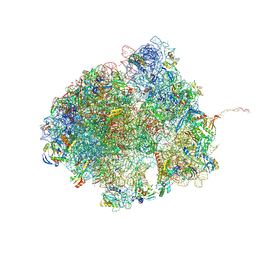 | | Bacillus subtilis MutS2-collided disome complex (collided 70S) | | 分子名称: | 16S rRNA (1533-MER), 23S RNA (2887-MER), 30S ribosomal protein S10, ... | | 著者 | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | 登録日 | 2023-11-16 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
