2O9V
 
 | |
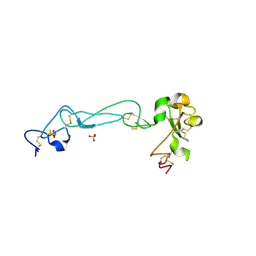
2AO7
 
 | | Adam10 Disintegrin and cysteine- rich domain | | Descriptor: | ADAM 10, SULFATE ION | | Authors: | Janes, P.W, Saha, N, Barton, W.A, Kolev, M.V, Wimmer-Kleikamp, S.H, Nievergall, E, Blobel, C.P, Himanen, J.-P, Lackmann, M, Nikolov, D.B. | | Deposit date: | 2005-08-12 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.
Cell(Cambridge,Mass.), 123, 2005
|
|
6NQD
 
 | |
1YE8
 
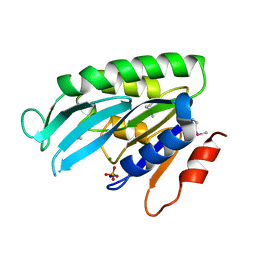 | | Crystal Structure of THEP1 from the hyperthermophile Aquifex aeolicus | | Descriptor: | Hypothetical UPF0334 kinase-like protein AQ_1292, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rossbach, M, Daumke, O, Klinger, C, Wittinghofer, A, Kaufmann, M. | | Deposit date: | 2004-12-28 | | Release date: | 2005-03-29 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of THEP1 from the hyperthermophile Aquifex aeolicus: a variation of the RecA fold
BMC Struct.Biol., 5, 2005
|
|
2OKW
 
 | |
2LB4
 
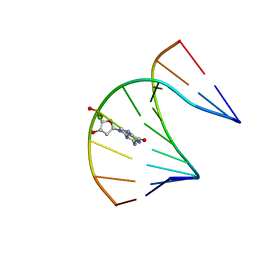 | | DNA / RNA Hybrid containing a central stereo specific Sp borano phosphate linkage | | Descriptor: | DNA_(5'-D(*AP*TP*GP*GP*TP*BGR*CP*TP*C)-3')_, RNA_(5'-R(*GP*AP*GP*CP*AP*CP*CP*AP*U)-3')_ | | Authors: | Johnson, C.N, Spring, A.M, Shaw, B.R, Germann, M.W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the RNase H1 Activity on Stereo Regular Borano Phosphonate DNA/RNA Hybrids.
Biochemistry, 50, 2011
|
|
1YA5
 
 | | Crystal structure of the titin domains z1z2 in complex with telethonin | | Descriptor: | N2B-TITIN ISOFORM, SULFATE ION, TELETHONIN | | Authors: | Pinotsis, N, Popov, A, Zou, P, Wilmanns, M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk
Nature, 439, 2006
|
|
2B6B
 
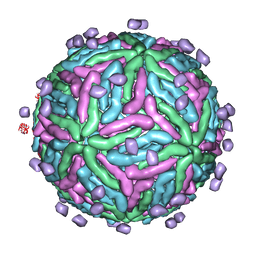 | | Cryo EM structure of Dengue complexed with CRD of DC-SIGN | | Descriptor: | CD209 antigen, envelope glycoprotein | | Authors: | Pokidysheva, E, Zhang, Y, Battisti, A.J, Bator-Kelly, C.M, Chipman, P.R, Gregorio, G, Hendrickson, W.A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-09-30 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN
Cell(Cambridge,Mass.), 124, 2006
|
|
2QZF
 
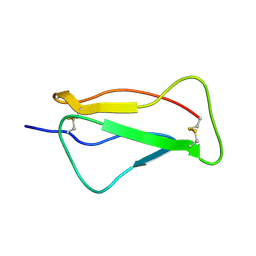 | | SCR1 of DAF from 1ojv fitted into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZH
 
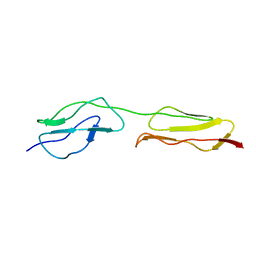 | | SCR2/3 of DAF from the NMR structure 1nwv fitted into a cryoEM reconstruction of CVB3-RD complexed with DAF | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2R29
 
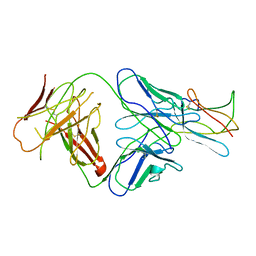 | | Neutralization of dengue virus by a serotype cross-reactive antibody elucidated by cryoelectron microscopy and x-ray crystallography | | Descriptor: | Envelope protein E, Heavy chain of Fab 1A1D-2, Light chain of Fab 1A1D-2 | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3L1P
 
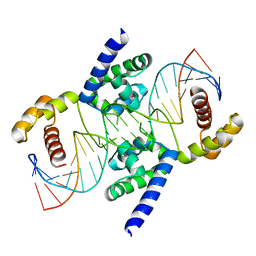 | | POU protein:DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP*TP*CP*AP*AP*AP*TP*GP*TP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP*CP*AP*AP*AP*TP*GP*GP*A)-3'), POU domain, ... | | Authors: | Vahokoski, J, Groves, M.R, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2009-12-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique Oct4 interface is crucial for reprogramming to pluripotency
Nat.Cell Biol., 15, 2013
|
|
2O0J
 
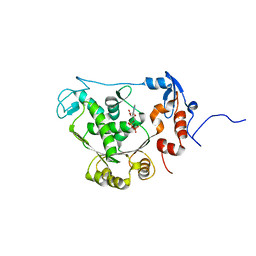 | | T4 gp17 ATPase domain mutant complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA packaging protein Gp17 | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2006-11-27 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Procapsids
MOL.CELL, 25, 2007
|
|
2LDB
 
 | |
2LAR
 
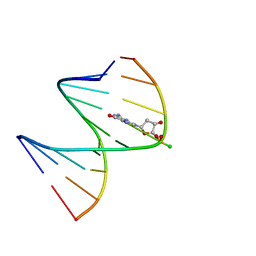 | | DNA / RNA Hybrid containing a central stereo specific Rp borano phosphate linkage | | Descriptor: | DNA_(5'-D(*AP*TP*GP*GP*TP*BGR*CP*TP*C)-3')_, RNA_(5'-R(*GP*AP*GP*CP*AP*CP*CP*AP*U)-3')_ | | Authors: | Johnson, C.N, Spring, A.M, Shaw, B.R, Germann, M.W. | | Deposit date: | 2011-03-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the RNase H1 activity on stereo regular borano phosphonate DNA/RNA hybrids.
Biochemistry, 50, 2011
|
|
2LDX
 
 | |
2LWG
 
 | |
2LWH
 
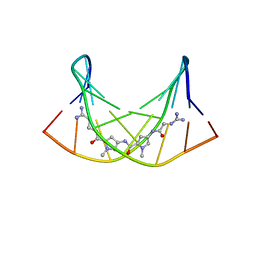 | | NMR Structure of the Self-Complementary 10 mer DNA Duplex 5'-GGATATATCC-3' in Complex with Netropsin | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*AP*TP*AP*TP*CP*C)-3'), NETROPSIN | | Authors: | Rettig, M, Germann, M.W, Wilson, W, Wang, S. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for sequence-dependent induced DNA bending.
Chembiochem, 14, 2013
|
|
1Z7Z
 
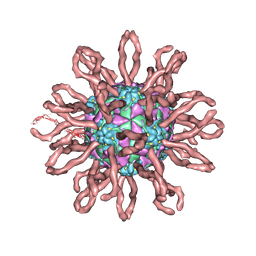 | | Cryo-em structure of human coxsackievirus A21 complexed with five domain icam-1kilifi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1, human coxsackievirus A21 | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
1A90
 
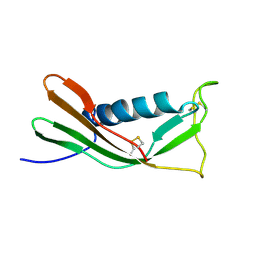 | | RECOMBINANT MUTANT CHICKEN EGG WHITE CYSTATIN, NMR, 31 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-04-14 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
2QZD
 
 | | Fitted structure of SCR4 of DAF into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2R15
 
 | |
2R69
 
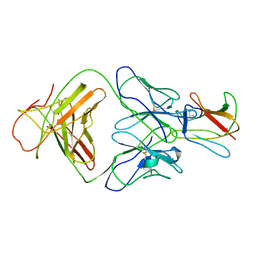 | | Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2C0M
 
 | |
2P73
 
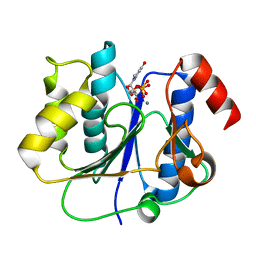 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
