1QDL
 
 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | Authors: | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1999-05-20 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2O0J
 
 | | T4 gp17 ATPase domain mutant complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA packaging protein Gp17 | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2006-11-27 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Procapsids
MOL.CELL, 25, 2007
|
|
4NZL
 
 | | Extracellular proteins of Staphylococcus aureus inhibit the neutrophil serine proteases | | Descriptor: | Neutrophil elastase, Uncharacterized protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stapels, D.A.C, von Koeckritz-Blickwede, M, Ramyar, K.X, Bischoff, M, Milder, F, Ruyken, M, Scheepmaker, L, McWhorter, W.J, Herrmann, M, van Kessel, K.P.M, Geisbrecht, B.V, Rooijakkers, S.H.M. | | Deposit date: | 2013-12-12 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1QNY
 
 | | X-ray refinement of D2O soaked crystal of concanavalin A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb(Gilboa), A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct Determination of the Positions of Deuterium Atoms of Bound Water in Concanavalin a by Neutron Laue Crystallography
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3HB2
 
 | | PrtC methionine mutants: M226I | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-04 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
2R15
 
 | |
5ADO
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - Light chain S35R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 | | Authors: | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | Deposit date: | 2015-08-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
5AF7
 
 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2RS3
 
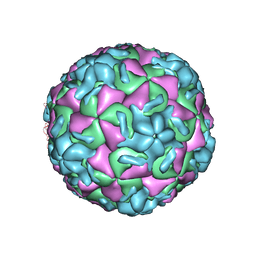 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-ETHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
6Y1L
 
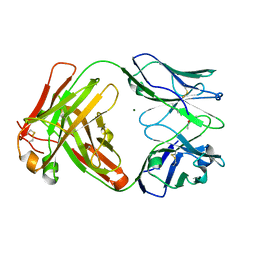 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4HR6
 
 | | Crystal structure of snake gourd (Trichosanthes anguina) seed lectin, a three chain homologue of type II RIPs | | Descriptor: | LECTIN, methyl alpha-D-galactopyranoside | | Authors: | Sharma, A, Pohlentz, G, Bobbili, K.B, Jeyaprakash, A.A, Chandran, T, Mormann, M, Swamy, M.J, Vijayan, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The sequence and structure of snake gourd (Trichosanthes anguina) seed lectin, a three-chain nontoxic homologue of type II RIPs.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6Y49
 
 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MID
 
 | |
6Y1K
 
 | | Crystal structure of the unmodified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | FAB A.17 L47R mutant Heavy Chain, FAB A.17 L47R mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MW9
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-3 antibody | | Descriptor: | E1, E2, EEEV-3 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWX
 
 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-69 Antibody | | Descriptor: | E1, E2, EEEV-69 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
2IC6
 
 | |
6Y1M
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47K mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light CHAIN | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7O2L
 
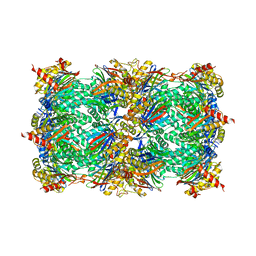 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
6MUI
 
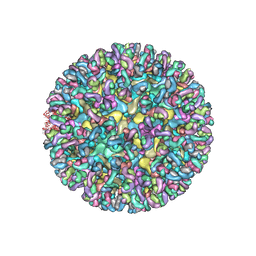 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-42 antibody | | Descriptor: | E1, E2, EEEV-42 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
1C8M
 
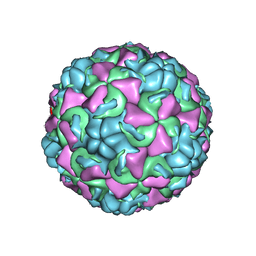 | | REFINED CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS 16 COMPLEXED WITH VP63843 (PLECONARIL), AN ANTI-PICORNAVIRAL DRUG CURRENTLY IN CLINICAL TRIALS | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, HUMAN RHINOVIRUS 16 COAT PROTEIN, ZINC ION | | Authors: | Chakravarty, S, Bator, C.M, Pevear, D.C, Diana, G.D, Rossmann, M.G. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE REFINED STRUCTURE OF A PICORNAVIRUS INHIBITOR CURRENTLY IN CLINICAL TRIALS, WHEN COMPLEXED WITH HUMAN RHINOVIRUS 16
to be published
|
|
6HQV
 
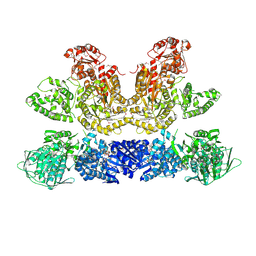 | | Pentafunctional AROM Complex from Chaetomium thermophilum | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Arora Verasto, H, Hartmann, M.D. | | Deposit date: | 2018-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture and functional dynamics of the pentafunctional AROM complex.
Nat.Chem.Biol., 16, 2020
|
|
6MWV
 
 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-58 Antibody | | Descriptor: | E1, E2, EEEV-58 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
4JV8
 
 | | The crystal structure of PDE6D in complex with rac-S1 | | Descriptor: | (6R)-6-(pyridin-2-yl)-5,6-dihydrobenzimidazo[1,2-c]quinazoline, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
