6CAA
 
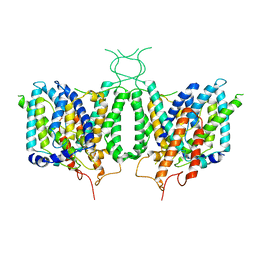 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
5OHM
 
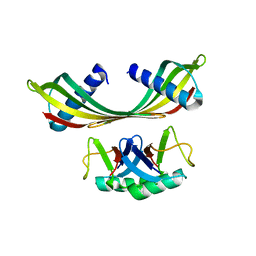 | | K33-specific affimer bound to K33 diUb | | Descriptor: | K33-specific affimer, POLYETHYLENE GLYCOL (N=34), Polyubiquitin-C | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5I4E
 
 | | Crystal Structure of Human Nonmuscle Myosin 2C motor domain | | Descriptor: | ADP ORTHOVANADATE, MAGNESIUM ION, Myosin-14,Alpha-actinin A | | Authors: | Chinthalapudi, K, Heissler, S.M, Preller, M, Sellers, J.R, Manstein, D.J. | | Deposit date: | 2016-02-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic insights into the active site and allosteric communication pathways in human nonmuscle myosin-2C.
Elife, 6, 2017
|
|
6GZT
 
 | |
5G5T
 
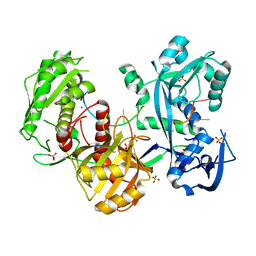 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii in complex with guide DNA | | Descriptor: | ARGONAUTE, GUIDE DNA, MAGNESIUM ION, ... | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
5G5S
 
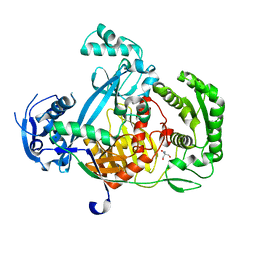 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGONAUTE, MAGNESIUM ION | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
7TVG
 
 | |
7TVF
 
 | |
6GLC
 
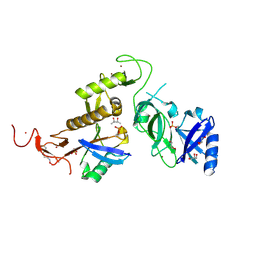 | | Structure of phospho-Parkin bound to phospho-ubiquitin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Gladkova, C, Maslen, S.L, Skehel, J.M, Komander, D. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of parkin activation by PINK1.
Nature, 559, 2018
|
|
5RL5
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-30 (Mpro-x3359) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-(ethylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
1FVQ
 
 | | SOLUTION STRUCTURE OF THE YEAST COPPER TRANSPORTER DOMAIN CCC2A IN THE APO AND CU(I) LOADED STATES | | Descriptor: | COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi Baffoni, S, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-09-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the yeast copper transporter domain Ccc2a in the apo and Cu(I)-loaded states.
J.Biol.Chem., 276, 2001
|
|
5RL3
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-39 (Mpro-x3117) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(oxan-4-yl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
4A7F
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 3) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
5RL2
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-26 (Mpro-x3115) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(2-methoxyethyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL0
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-2 (Mpro-x3110) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL4
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-3 (Mpro-x3124) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-(methylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5OO3
 
 | |
5OSJ
 
 | | Cdk2(WT) with covalent adduct at C177 | | Descriptor: | Cyclin-dependent kinase 2, ~{tert}-butyl 4-propanoyl-2,3-dihydroquinoxaline-1-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5OSM
 
 | | Cdk2(F80C, C177A) with covalent adduct at C80 | | Descriptor: | Cyclin-dependent kinase 2, methyl 1-propanoyl-3,4-dihydro-2~{H}-quinoline-6-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4AK9
 
 | |
7RS6
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to GMPCPP-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2024
|
|
7RS5
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to Taxol-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2021
|
|
7S6O
 
 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7S3T
 
 | | NzeB Diketopiperazine Dimerase Mutant: Q68I-G87A-A89G-I90V | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Harris, N.R, Shende, V.V, Sanders, J.N, Newmister, S.A, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 2023
|
|
