5KTY
 
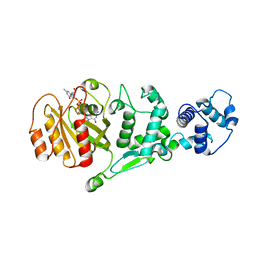 | | hMiro EF hand and cGTPase domains, GDP and Ca2+ bound state | | 分子名称: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | 登録日 | 2016-07-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.522 Å) | | 主引用文献 | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
3HIZ
 
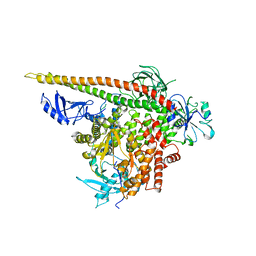 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | 登録日 | 2009-05-20 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HHM
 
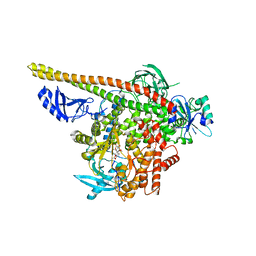 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | 分子名称: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | 著者 | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | 登録日 | 2009-05-15 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5KSY
 
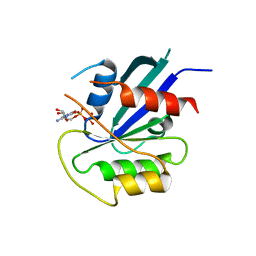 | | hMiro1 C-domain GDP Complex P41212 Crystal Form | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | 著者 | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | 登録日 | 2016-07-10 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.482 Å) | | 主引用文献 | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KTZ
 
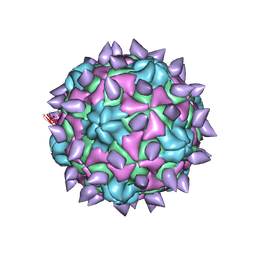 | | expanded poliovirus in complex with VHH 12B | | 分子名称: | Genome polyprotein, VHH 12B, VP1, ... | | 著者 | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5K21
 
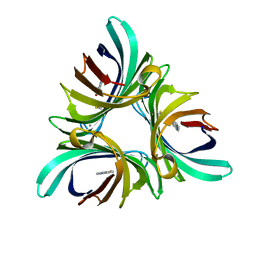 | | Pyocyanin demethylase | | 分子名称: | CALCIUM ION, Pyocyanin demethylase, phenazin-1-ol | | 著者 | Costa, K.C, Glasser, N.R, Newman, D.K. | | 登録日 | 2016-05-18 | | 公開日 | 2016-12-07 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms.
Science, 355, 2017
|
|
8EEQ
 
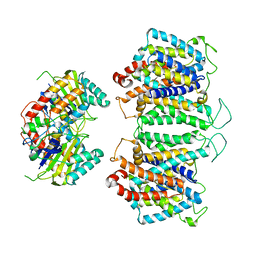 | | CryoEM structures of bAE1 captured in multiple states. | | 分子名称: | Anion exchange protein | | 著者 | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | 登録日 | 2022-09-07 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8E34
 
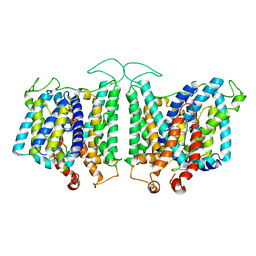 | | CryoEM structures of bAE1 captured in multiple states | | 分子名称: | Anion exchange protein | | 著者 | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | 登録日 | 2022-08-16 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8EBZ
 
 | |
7AAP
 
 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | 分子名称: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | 著者 | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | 登録日 | 2020-09-04 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3I2M
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi s Sarcoma-Associated Herpesvirus | | 分子名称: | ORF59 | | 著者 | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | 登録日 | 2009-06-29 | | 公開日 | 2010-05-12 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
3HSL
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi's Sarcoma-Associated Herpesvirus | | 分子名称: | ORF59 | | 著者 | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | 登録日 | 2009-06-10 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
3KB2
 
 | | Crystal Structure of YorR protein in complex with phosphorylated GDP from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR256 | | 分子名称: | GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION, SPBc2 prophage-derived uncharacterized protein yorR | | 著者 | Forouhar, F, Friedman, D, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Ma, L, Ho, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-10-20 | | 公開日 | 2009-10-27 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target SR256
To be Published
|
|
1Z1B
 
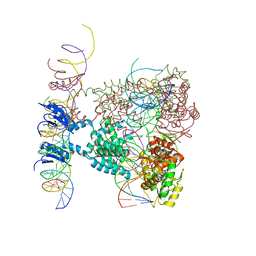 | | Crystal structure of a lambda integrase dimer bound to a COC' core site | | 分子名称: | 26-MER DNA, 29-MER DNA, 5'-D(*CP*T*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', ... | | 著者 | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | 登録日 | 2005-03-03 | | 公開日 | 2005-06-28 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
1Z19
 
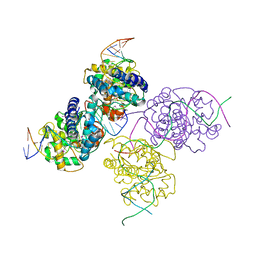 | | Crystal structure of a lambda integrase(75-356) dimer bound to a COC' core site | | 分子名称: | 33-MER, 5'-D(*CP*TP*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*AP*TP*AP*CP*TP*AP*AP*GP*TP*TP*GP*GP*CP*AP*TP*TP*A)-3', ... | | 著者 | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | 登録日 | 2005-03-03 | | 公開日 | 2005-06-28 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
1Z1G
 
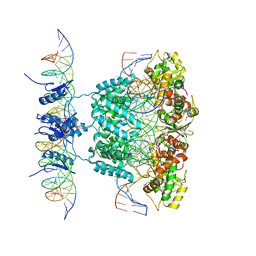 | | Crystal structure of a lambda integrase tetramer bound to a Holliday junction | | 分子名称: | 25-MER, 29-MER, 5'-D(*AP*CP*AP*GP*GP*TP*CP*AP*CP*TP*AP*TP*CP*AP*GP*TP*CP*AP*AP*AP*AP*TP*AP*CP*C)-3', ... | | 著者 | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | 登録日 | 2005-03-03 | | 公開日 | 2005-06-28 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
4WXU
 
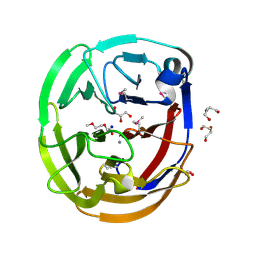 | | Crystal Structure of the Selenomthionine Incorporated Myocilin Olfactomedin Domain E396D Variant. | | 分子名称: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | 登録日 | 2014-11-14 | | 公開日 | 2015-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
4WXS
 
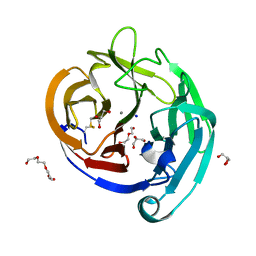 | | Crystal Structure of the E396D SNP Variant of the Myocilin Olfactomedin Domain | | 分子名称: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | 登録日 | 2014-11-14 | | 公開日 | 2015-04-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
3J8F
 
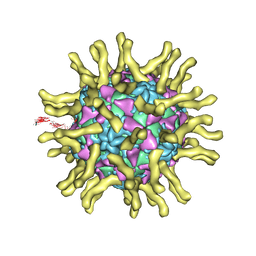 | | Cryo-EM reconstruction of poliovirus-receptor complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | 著者 | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | 登録日 | 2014-10-20 | | 公開日 | 2015-02-11 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3J9F
 
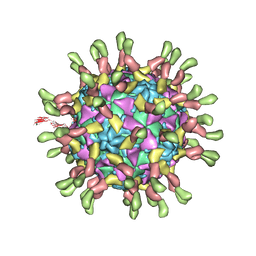 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | 著者 | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | 登録日 | 2015-01-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
1AS3
 
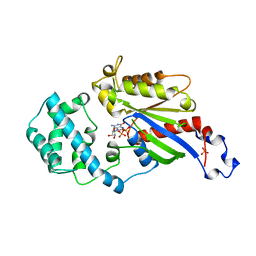 | | GDP BOUND G42V GIA1 | | 分子名称: | GIA1, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | 著者 | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | 登録日 | 1997-08-11 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1AS0
 
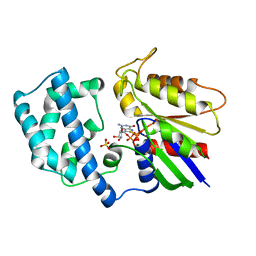 | | GTP-GAMMA-S BOUND G42V GIA1 | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GIA1, MAGNESIUM ION, ... | | 著者 | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | 登録日 | 1997-08-11 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1AS2
 
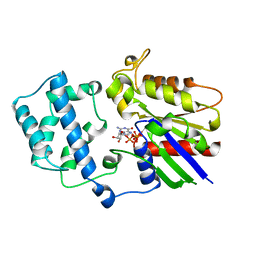 | | GDP+PI BOUND G42V GIA1 | | 分子名称: | GIA1, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | 著者 | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | 登録日 | 1997-08-11 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1CQU
 
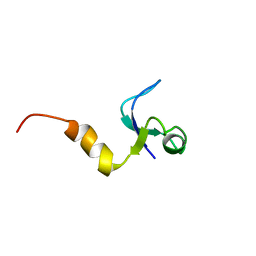 | |
7ABK
 
 | | Helical structure of PspA | | 分子名称: | Chloroplast membrane-associated 30 kD protein | | 著者 | Junglas, B, Huber, S.T, Mann, D, Heidler, T, Clarke, M, Schneider, D, Sachse, C. | | 登録日 | 2020-09-07 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | PspA adopts an ESCRT-III-like fold and remodels bacterial membranes.
Cell, 184, 2021
|
|
