1PO1
 
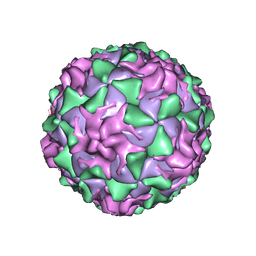 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R80633, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7O9J
 
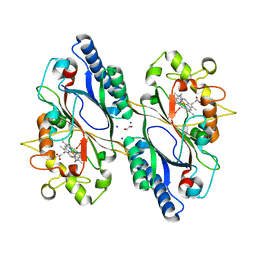 | | Crystal structure of DyP-type peroxidase from Dictyostelium discoideum in complex with an activated form of oxygen | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
7O9L
 
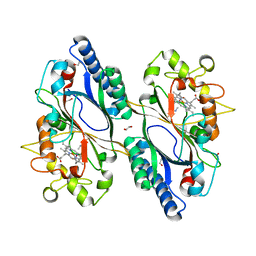 | | Dictyostelium discoideum dye decolorizing peroxidase DyPA in complex with cyanide. | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, DyPA, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
6HEJ
 
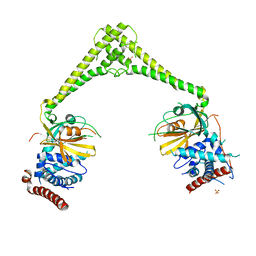 | | Structure of human USP28 | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
2PAN
 
 | | Crystal structure of E. coli glyoxylate carboligase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kaplun, A, Chipman, D.M, Barak, Z, Vyazmensky, M, Shaanan, B. | | Deposit date: | 2007-03-27 | | Release date: | 2008-01-01 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes.
Nat.Chem.Biol., 4, 2008
|
|
6PVF
 
 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVH
 
 | | Crystal structure of PhqK in complex with paraherquamide K | | Descriptor: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVI
 
 | | Crystal structure of PhqK in complex with paraherquamide L | | Descriptor: | (8aS,13S,13aR,14aS)-4,4,13,15,15-pentamethyl-12,13,14,14a,15,16-hexahydro-4H,8H,9H,11H-8a,13a-(epiminomethano)[1,4]dioxepino[2,3-a]indolizino[6,7-h]carbazol-17-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVJ
 
 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
7FDS
 
 | | High resolution crystal structure of LpqH from Mycobacterium tuberculosis | | Descriptor: | Lipoprotein LpqH | | Authors: | Kundapura, S.V, Chatterjee, S, Samanta, D, Ramagopal, U.A. | | Deposit date: | 2021-07-17 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | High-resolution crystal structure of LpqH, an immunomodulatory surface lipoprotein of Mycobacterium tuberculosis reveals a distinct fold and a conserved cleft on its surface.
Int.J.Biol.Macromol., 210, 2022
|
|
3NS8
 
 | |
1POV
 
 | |
2H7Y
 
 | | Pikromycin Thioesterase with covalent affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
6HEK
 
 | | Structure of human USP28 bound to Ubiquitin-PA | | Descriptor: | CHLORIDE ION, Polyubiquitin-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
3OKV
 
 | | Human Carbonic Anhydrase II A65S, N67Q (CA IX mimic) bound with 2-Ethylestrone 3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
2I3Q
 
 | | Q44V mutant of Homing Endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*AP*CP*TP*CP*AP*CP*GP*TP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*AP*CP*GP*TP*GP*AP*GP*TP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Rosen, L, Sussman, D. | | Deposit date: | 2006-08-20 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homing endonuclease I-CreI derivatives with novel DNA target specificities.
Nucleic Acids Res., 34, 2006
|
|
3O7V
 
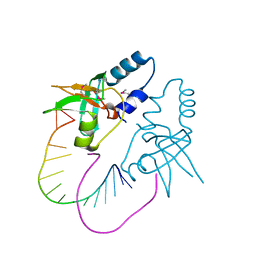 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1PVC
 
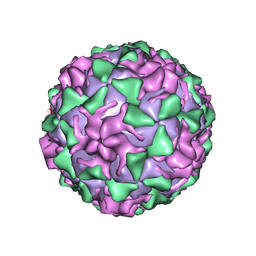 | |
3O6E
 
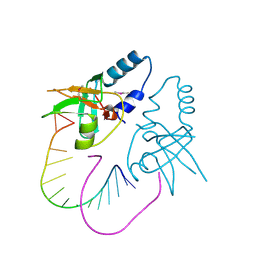 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6GZS
 
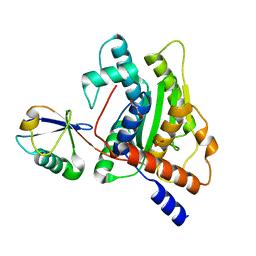 | |
1LEV
 
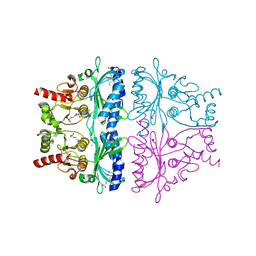 | | PORCINE KIDNEY FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH AN AMP-SITE INHIBITOR | | Descriptor: | 3-(2-CARBOXY-ETHYL)-4,6-DICHLORO-1H-INDOLE-2-CARBOXYLIC ACID, 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, ... | | Authors: | Wright, S.W, Carlo, A.A, Danley, D.E, Hageman, D.L, Karam, G.A, Mansour, M.N, McClure, L.D, Pandit, J, Schulte, G.K, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-04-10 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3-(2-carboxyethyl)-4,6-dichloro-1H-indole-2-carboxylic acid: an allosteric inhibitor of fructose-1,6-bisphosphatase at the AMP site.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
7F9F
 
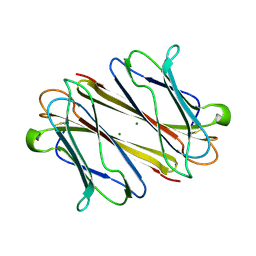 | | Thrombocorticin | | Descriptor: | MAGNESIUM ION, Thrombocorticin | | Authors: | Kageyama, H, Onodera, K, Sakai, R, Tanaka, Y, Freymann, D.M. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | A marine sponge-derived lectin reveals hidden pathway for thrombopoietin receptor activation.
Nat Commun, 13, 2022
|
|
1KZ8
 
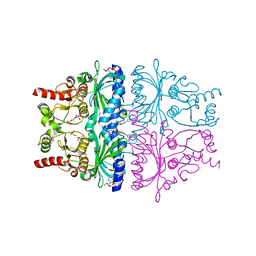 | | CRYSTAL STRUCTURE OF PORCINE FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH A NOVEL ALLOSTERIC-SITE INHIBITOR | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Wright, S.W, Carlo, A.A, Carty, M.D, Danley, D.E, Hageman, D.L, Karam, G.A, Levy, C.B, Mansour, M.N, Mathiowetz, A.M, McClure, L.D, Nestor, N.B, McPherson, R.K, Pandit, J, Pustilnik, L.R, Schulte, G.K, Soeller, W.C, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-02-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ANILINOQUINAZOLINE INHIBITORS OF FRUCTOSE 1,6-BISPHOSPHATASE BIND AT A NOVEL ALLOSTERIC SITE: SYNTHESIS, IN VITRO CHARACTERIZATION, AND X-RAY CRYSTALLOGRAPHY
J.MED.CHEM., 45, 2002
|
|
1JWY
 
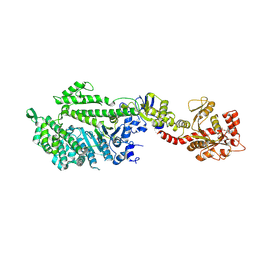 | | CRYSTAL STRUCTURE OF THE DYNAMIN A GTPASE DOMAIN COMPLEXED WITH GDP, DETERMINED AS MYOSIN FUSION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Niemann, H.H, Knetsch, M.L.W, Scherer, A, Manstein, D.J, Kull, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms.
EMBO J., 20, 2001
|
|
2HFK
 
 | | Pikromycin thioesterase in complex with product 10-deoxymethynolide | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
