1XOK
 
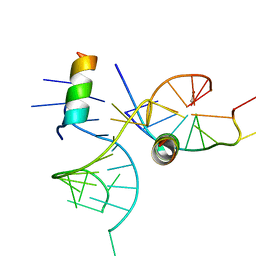 | | crystal structure of alfalfa mosaic virus RNA 3'UTR in complex with coat protein N terminal peptide | | Descriptor: | BROMIDE ION, Coat protein, alfalfa mosaic virus RNA 3' UTR | | Authors: | Guogas, L.M, Filman, D.J, Hogle, J.M, Gehrke, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cofolding organizes alfalfa mosaic virus RNA and coat protein for replication.
Science, 306, 2004
|
|
4RGF
 
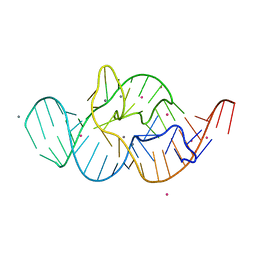 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
6XHA
 
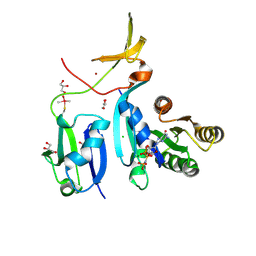 | | Crystal Structure of KRAS-G12V (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoform 2B of GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
3NF5
 
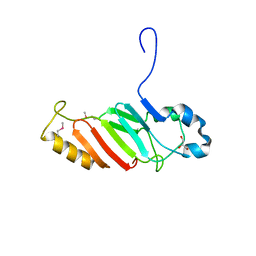 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
1YTR
 
 | | NMR structure of plantaricin a in dpc micelles, 20 structures | | Descriptor: | Bacteriocin plantaricin A | | Authors: | Kristiansen, P.E, Fimland, G, Mantzilas, D, Nissen-Meyer, J. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin A
J.Biol.Chem., 280, 2005
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
1CC8
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, MERCURY (II) ION, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
1XV8
 
 | | Crystal Structure of Human Salivary Alpha-Amylase Dimer | | Descriptor: | Alpha-amylase, CALCIUM ION, CHLORIDE ION | | Authors: | Fisher, S.Z, Govindasamy, L, Tu, C.K, Silverman, D.N, Rajaniemi, H, McKenna, R. | | Deposit date: | 2004-10-27 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Salivary Alpha-Amylase Dimer
To be Published
|
|
4S14
 
 | | Crystal structure of the orphan nuclear receptor RORgamma ligand-binding domain in complex with 4alpha-caboxyl, 4beta-methyl-zymosterol (4ACD8) | | Descriptor: | (3beta,4alpha,5beta,14beta)-3-hydroxy-4-methylcholesta-8,24-diene-4-carboxylic acid, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Huang, P, Santori, F.R, Littman, D.R, Rastinejad, F. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (3.542 Å) | | Cite: | Identification of Natural ROR gamma Ligands that Regulate the Development of Lymphoid Cells.
Cell Metab, 21, 2015
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
6RV3
 
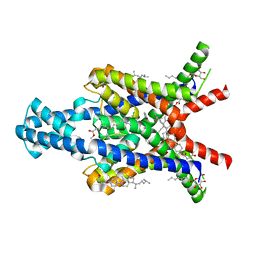 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
3C3T
 
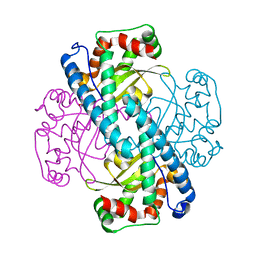 | | Role of a Glutamate Bridge Spanning the Dimeric Interface of Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] | | Authors: | Quint, P.S, Domsic, J.F, Cabelli, D.E, McKenna, R, Silverman, D.N. | | Deposit date: | 2008-01-28 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of a glutamate bridge spanning the dimeric interface of human manganese superoxide dismutase.
Biochemistry, 47, 2008
|
|
3BJ8
 
 | |
1A56
 
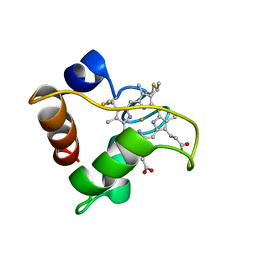 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERRICYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITH EXPLICIT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERRICYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-02-20 | | Release date: | 1998-10-21 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
1PLG
 
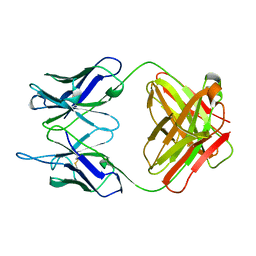 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
5A3G
 
 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3OT9
 
 | | Phosphopentomutase from Bacillus cereus bound to glucose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Phalen, V, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-09-10 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3C3S
 
 | | Role of a Glutamate Bridge Spanning the Dimeric Interface of Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] | | Authors: | Quint, P.S, Domsic, J.F, Cabelli, D.E, McKenna, R, Silverman, D.N. | | Deposit date: | 2008-01-28 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of a glutamate bridge spanning the dimeric interface of human manganese superoxide dismutase.
Biochemistry, 47, 2008
|
|
2ADP
 
 | | Nitrated Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, Superoxide dismutase [Mn] | | Authors: | Quint, P, Reutzel, R, Mikulski, R, McKenna, R, Silverman, D.N. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of nitrated human manganese superoxide dismutase: mechanism of inactivation.
FREE RADIC.BIOL.MED., 40, 2006
|
|
2ADQ
 
 | | Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, Superoxide dismutase [Mn] | | Authors: | Quint, P, Reutzel, R, Mikulski, R, McKenna, R, Silverman, D.N. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of nitrated human manganese superoxide dismutase: mechanism of inactivation.
FREE RADIC.BIOL.MED., 40, 2006
|
|
5HAF
 
 | |
2Y8I
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
4OUD
 
 | | Engineered tyrosyl-tRNA synthetase with the nonstandard amino acid L-4,4-biphenylalanine | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Takeuchi, R, Mandell, D.J, Lajoie, M.J, Church, G.M, Stoddard, B.L. | | Deposit date: | 2014-02-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biocontainment of genetically modified organisms by synthetic protein design.
Nature, 518, 2015
|
|
4OYK
 
 | |
