8ED8
 
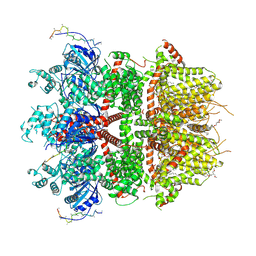 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2 and PregS, state 1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED9
 
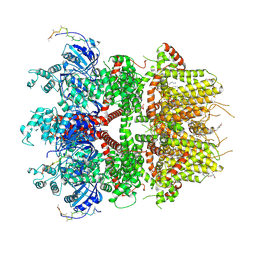 | | cryo-EM structure of TRPM3 ion channel in the presence with PIP2 and PregS, state 2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8GHF
 
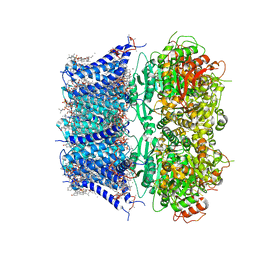 | | cryo-EM structure of hSlo1 in plasma membrane vesicles | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Tao, X, Zhao, C, MacKinnon, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Membrane protein isolation and structure determination in cell-derived membrane vesicles.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4KFM
 
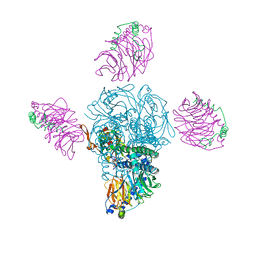 | |
7S5Y
 
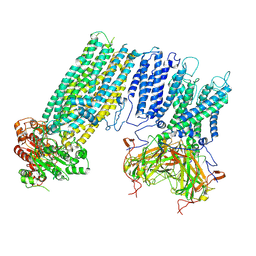 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S5Z
 
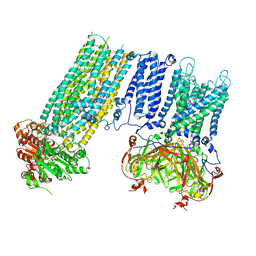 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4I9W
 
 | |
1OTS
 
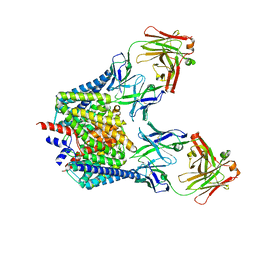 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTU
 
 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
8ED7
 
 | | cryo-EM structure of TRPM3 ion channel in apo state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Transient receptor potential cation channel, subfamily M, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8EMX
 
 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on lipid nanodiscs | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EMV
 
 | | Phospholipase C beta 3 (PLCb3) in solution | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EMW
 
 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on liposomes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DDU
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDS
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDT
 
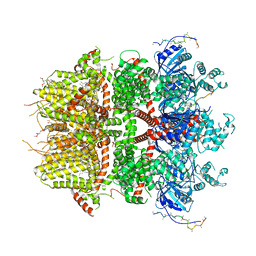 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDR
 
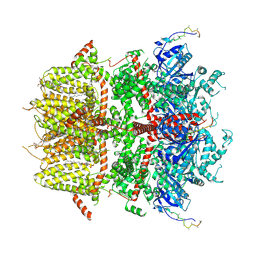 | | cryo-EM structure of TRPM3 ion channel in the absence of PIP2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDX
 
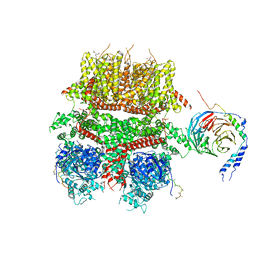 | | cryo-EM structure of TRPM3 ion channel in complex with Gbg in the presence of PIP2, tethered by ALFA-nanobody | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDV
 
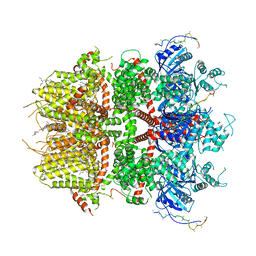 | | Cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
6PBY
 
 | |
6PBX
 
 | |
8GHG
 
 | |
8GH9
 
 | |
1ORQ
 
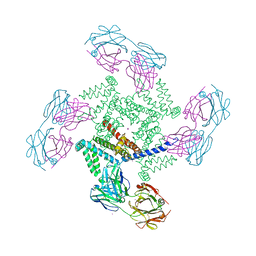 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
