7T2Q
 
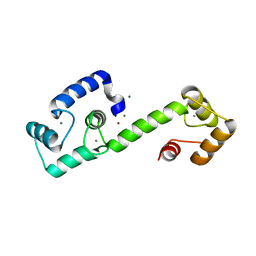 | | PEGylated Calmodulin-1 (K148U) | | Descriptor: | CALCIUM ION, Calmodulin-1, MAGNESIUM ION, ... | | Authors: | Mackay, J.P, Payne, R.J, Patel, K, Dowman, L.J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine.
Nat Commun, 13, 2022
|
|
7TO9
 
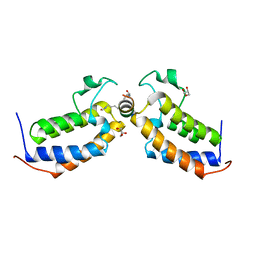 | |
7TO7
 
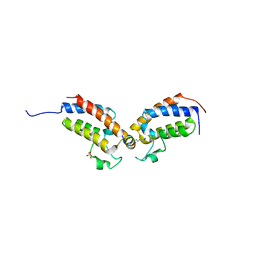 | |
2HI3
 
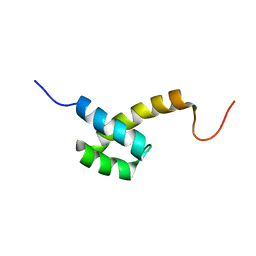 | | Solution structure of the homeodomain-only protein HOP | | Descriptor: | Homeodomain-only protein | | Authors: | Mackay, J.P, Kook, H, Epstein, J.A, Simpson, R.J, Yung, W.W. | | Deposit date: | 2006-06-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the structure and function of the transcriptional coregulator HOP
Biochemistry, 45, 2006
|
|
2MPL
 
 | |
8DNQ
 
 | | BRD2-BD1 in complex with cyclic peptide 2.2B | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 2.2B, GLYCEROL | | Authors: | Patel, K, Franck, C, Mackay, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
6PTL
 
 | |
8CV5
 
 | | Peptide 4.2B in complex with BRD3.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Bromodomain-containing protein 3, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV7
 
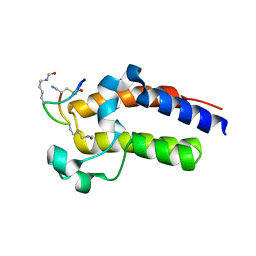 | | Peptide 2.2E in complex with BRD2-BD2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Isoform 3 of Bromodomain-containing protein 2, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV6
 
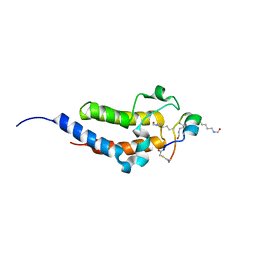 | | Peptide 4.2B in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV4
 
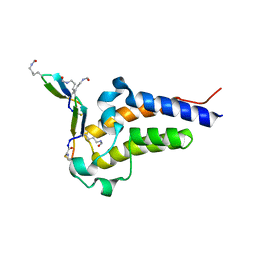 | | Peptide 4.2C in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
7JX7
 
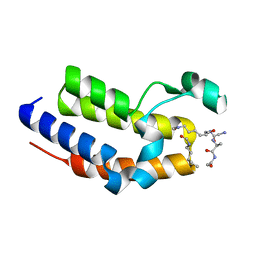 | | BRD2-BD2 in complex with a diacetylated-H2A.Z peptide | | Descriptor: | Bromodomain-containing protein 2, Diacetylated-H2A.Z peptide | | Authors: | Patel, K, Low, J.K.K, Mackay, J.P, Solomon, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The bromodomains of BET family proteins can recognize diacetylated histone H2A.Z.
Protein Sci., 30, 2021
|
|
7L4Z
 
 | | Structure of SARS-CoV-2 spike RBD in complex with cyclic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2, Spike protein S1 | | Authors: | Christie, M, Mackay, J.P, Passioura, T, Payne, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Discovery of Cyclic Peptide Ligands to the SARS-CoV-2 Spike Protein Using mRNA Display.
Acs Cent.Sci., 7, 2021
|
|
8D4Y
 
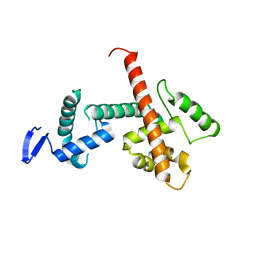 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
3G9Y
 
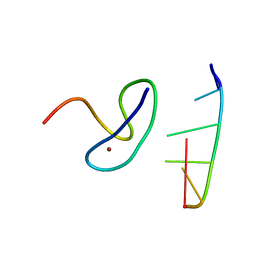 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7UQT
 
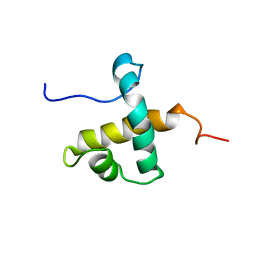 | | Solution NMR structure of hexahistidine tagged QseM (6H-QseM) | | Descriptor: | Quorum sensing master protein | | Authors: | Hall, D.A, Solomon, P.D, Bond, C.S, Ramsay, J.P, Mackay, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | DUF2285 is a novel helix-turn-helix domain variant that orchestrates both activation and antiactivation of conjugative element transfer in proteobacteria.
Nucleic Acids Res., 51, 2023
|
|
3OVU
 
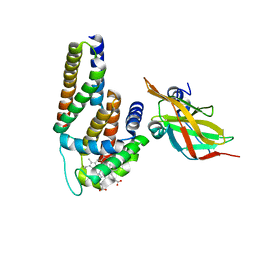 | | Crystal Structure of Human Alpha-Haemoglobin Complexed with AHSP and the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Alpha-hemoglobin-stabilizing protein, Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Krishna Kumar, K, Caradoc-Davies, T.T, Langley, D.B, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A new haem pocket structure in alpha-haemoglobin
To be Published
|
|
6OM5
 
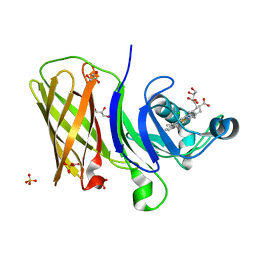 | | Structure of a haemophore from Haemophilus haemolyticus | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Torrado, M, Walshe, J.L, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A heme-binding protein produced by Haemophilus haemolyticus inhibits non-typeable Haemophilus influenzae.
Mol.Microbiol., 113, 2020
|
|
7TO8
 
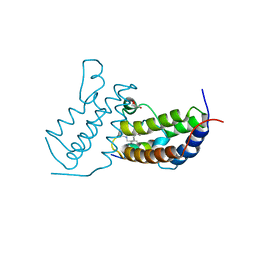 | |
7TOA
 
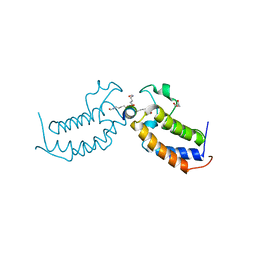 | |
3UK3
 
 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rediscovering DNA recognition by classical Zinc Fingers.
To be Published
|
|
4IS1
 
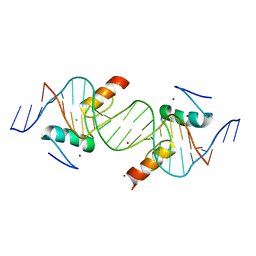 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217.
J.Biol.Chem., 288, 2013
|
|
6U4A
 
 | | BRD3-BD1 in complex with the cyclic peptide 3.1_3 | | Descriptor: | Bromodomain-containing protein 3, GLYCEROL, SULFATE ION, ... | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U6K
 
 | | BRD4-BD1 in complex with the cyclic peptide 3.1_3 | | Descriptor: | Bromodomain-containing protein 4, cyclic peptide 3.1_3 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U71
 
 | | BRD2-BD2 in complex with the cyclic peptide 3.1_3 | | Descriptor: | Bromodomain-containing protein 2, cyclic peptide 3.1_3 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
