4INT
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU122 | | Descriptor: | HMB-Val-Ser-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
8FMQ
 
 | |
6PTS
 
 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FMN
 
 | |
7D68
 
 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
7MLF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
2RQL
 
 | |
8FMR
 
 | |
8FMT
 
 | |
8FMO
 
 | |
7MLG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C63 | | Descriptor: | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide, 3C-like proteinase | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
4INU
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU112 | | Descriptor: | N3Phe-Phe(4-NH2CH2)-Leu-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
6SRP
 
 | |
6CIG
 
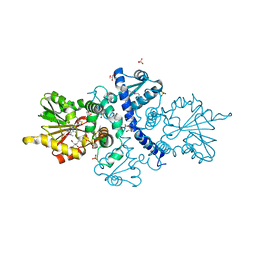 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
6SP4
 
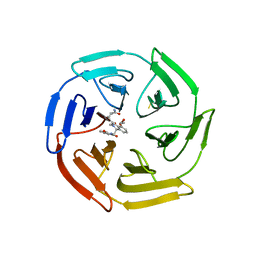 | | KEAP1 IN COMPLEX WITH COMPOUND 23 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclobutane-1-carboxamide, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
8FMS
 
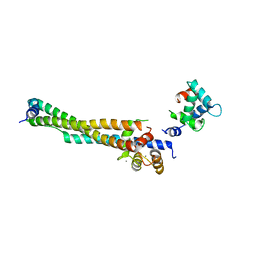 | |
6PX1
 
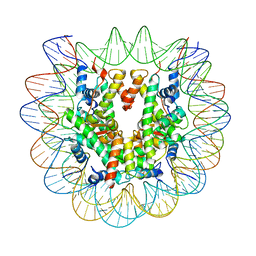 | | Set2 bound to nucleosome | | Descriptor: | DNA (149-MER), Histone H2B 1.1, Histone H3, ... | | Authors: | Halic, M, Bilokapic, S. | | Deposit date: | 2019-07-24 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
6SQW
 
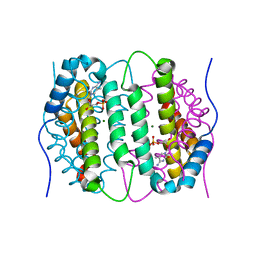 | | Mouse dCTPase in complex with 5-Me-dCMP | | Descriptor: | 5-METHYL-2'-DEOXY-CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
6MII
 
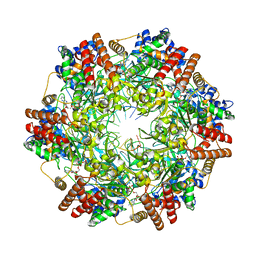 | | Crystal structure of minichromosome maintenance protein MCM/DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Meagher, M, Epling, L.B. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | DNA translocation mechanism of the MCM complex and implications for replication initiation.
Nat Commun, 10, 2019
|
|
2JCN
 
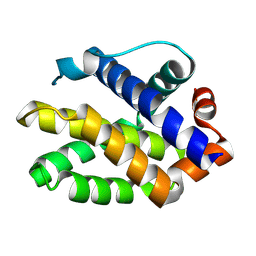 | | The crystal structure of BAK1 - a mitochondrial apoptosis regulator | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, SULFATE ION | | Authors: | Moche, M, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Lundgren, S, Nilsson, M.E, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-27 | | Release date: | 2007-01-04 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Bak1 - an Apoptosis Trigger in the Mitochondrial Outer Membrane
To be Published
|
|
6QF7
 
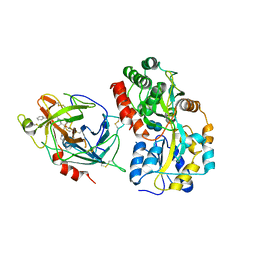 | | Crystal structures of the recombinant beta-Factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, ... | | Authors: | Pathak, M, Mannal, R, Li, C, Bubacarr, G.K, Badraldin, K.H, Belviso, B.D, Camila, R.B, Dreveny, I, Fischer, P.M, Dekker, L.V, Oliva, M.L.V, Emsley, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6SR4
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 112 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6SRB
 
 | | Crystal structure of glutathione transferase Omega 3C from Trametes versicolor | | Descriptor: | GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
