7A9W
 
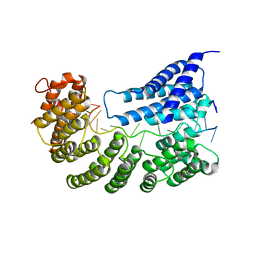 | | Structure of yeast Rmd9p in complex with 20nt target RNA | | Descriptor: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | Authors: | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-04-07 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SFB
 
 | |
6RX1
 
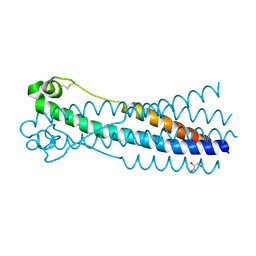 | | Crystal structure of human syncytin 1 in post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Syncytin-1 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
7A9X
 
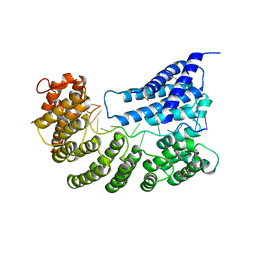 | | Structure of yeast Rmd9p in complex with 16nt target RNA | | Descriptor: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | Authors: | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SFI
 
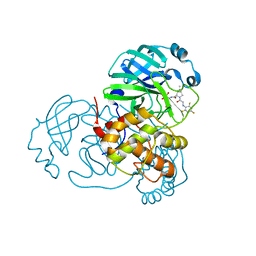 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML104 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-03 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | Descriptor: | Muramoyltetrapeptide carboxypeptidase | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
6MFP
 
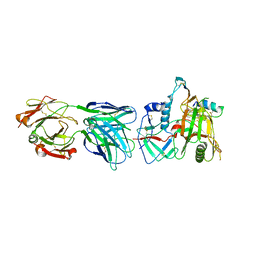 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
7SB6
 
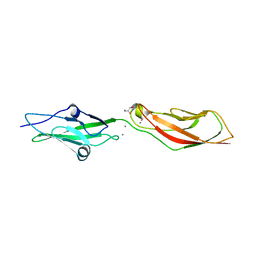 | | Crystal Structure of Ancestral Mammalian Cadherin-23 EC1-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin 23, ... | | Authors: | Nisler, C.R, Narui, Y, Sotomayor, M. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Interpreting the Evolutionary Echoes of a Protein Complex Essential for Inner-Ear Mechanosensation.
Mol.Biol.Evol., 40, 2023
|
|
7SH4
 
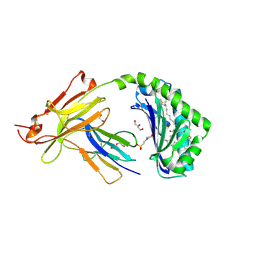 | | CD1a-phosphatidylglycerol binary structure | | Descriptor: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wegrecki, M, Rossjohn, J. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a.
Nat.Immunol., 24, 2023
|
|
7A5U
 
 | |
1USR
 
 | | Newcastle disease virus hemagglutinin-neuraminidase: Evidence for a second sialic acid binding site and implications for fusion | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zaitsev, V, Von Itzstein, M, Groves, D, Kiefel, M, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2003-11-28 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Second Sialic Acid Binding Site in Newcastle Disease Virus Hemagglutinin-Neuraminidase: Implications for Fusion
J.Virol., 78, 2004
|
|
6LL7
 
 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mn-activated form | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, MANGANESE (II) ION | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
1HRU
 
 | | THE STRUCTURE OF THE YRDC GENE PRODUCT FROM E.COLI | | Descriptor: | PHOSPHATE ION, YRDC GENE PRODUCT | | Authors: | Teplova, M, Tereshko, V, Sanishvili, R, Joachimiak, A, Bushueva, T, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding.
Protein Sci., 9, 2000
|
|
6VH7
 
 | | Doublet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
1UWS
 
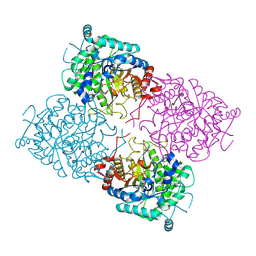 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-glucose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
6UOS
 
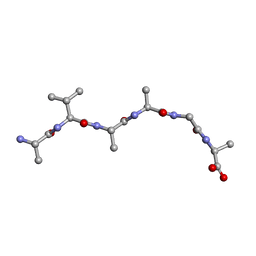 | |
6UOW
 
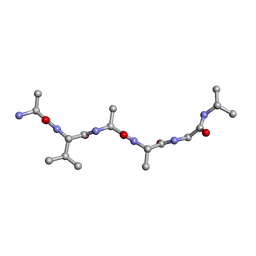 | |
5AQD
 
 | | Crystal structure of Phormidium Phycoerythrin at pH 8.5 | | Descriptor: | GLYCEROL, PHYCOERYTHRIN ALPHA SUBUNIT, PHYCOERYTHRIN BETA SUBUNIT, ... | | Authors: | Kumar, V, Sharma, M, Sonani, R.R, Gupta, G.D, Madamwar, D. | | Deposit date: | 2015-09-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
6V06
 
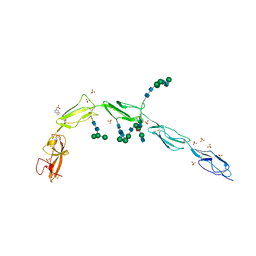 | | Crystal structure of Beta-2 glycoprotein I purified from plasma (pB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-glycoprotein 1, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V09
 
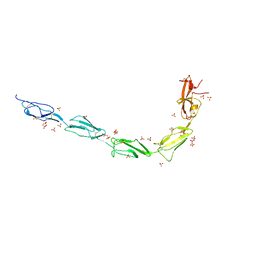 | | Crystal structure of human recombinant Beta-2 glycoprotein I short tag (ST-B2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6UPT
 
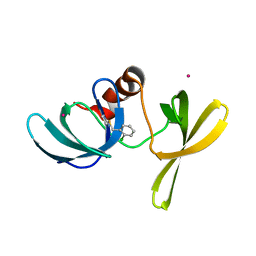 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706 | | Descriptor: | 2-((2-chlorobenzyl)thio)-4,5-dihydro-1H-imidazole, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706
to be published
|
|
6V0V
 
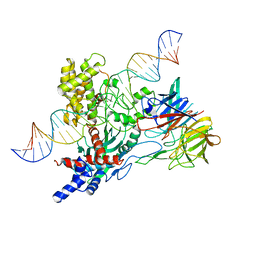 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1V4B
 
 | |
6M9D
 
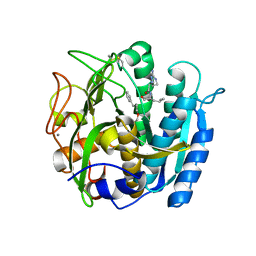 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Chymostatin | | Descriptor: | CALCIUM ION, Chymostatin A, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
