1IOK
 
 | | CRYSTAL STRUCTURE OF CHAPERONIN-60 FROM PARACOCCUS DENITRIFICANS | | Descriptor: | CHAPERONIN 60 | | Authors: | Fukami, T.A, Yohda, M, Taguchi, H, Yoshida, M, Miki, K. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of chaperonin-60 from Paracoccus denitrificans.
J.Mol.Biol., 312, 2001
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1IX4
 
 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Carbon Monoxide | | Descriptor: | CARBON MONOXIDE, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1IRR
 
 | | Solution structure of paralytic peptide of the silkworm, Bombyx mori | | Descriptor: | paralytic peptide | | Authors: | Miura, K, Kamimura, M, Aizawa, T, Kiuchi, M, Hayakawa, Y, Mizuguchi, M, Kawano, K. | | Deposit date: | 2001-10-23 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of paralytic peptide of silkworm, Bombyx mori
peptides, 23, 2002
|
|
8QI7
 
 | |
5F8Z
 
 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein LIGHT CHAIN, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1
To Be Published
|
|
1IX5
 
 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
8Q92
 
 | | P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
8Q96
 
 | | P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau, Unknown protein | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
6TA5
 
 | | OprM-MexA complex from the MexAB-OprM Pseudomonas aeruginosa whole assembly reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
5D5A
 
 | | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Liu, X, Kobilka, B, Kay Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4826 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | Descriptor: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | Authors: | Shi, R, Picard, M.-E, Manenda, M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
5M3M
 
 | | Free monomeric RNA polymerase I at 4.0A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
6TDO
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket. | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
6TDQ
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket and one GM dipeptide in the F pocket. | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDS
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule without peptide bound after NaCl wash | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6DR8
 
 | | Metallo-beta-lactamase from Cronobacter sakazakii (Enterobacter sakazakii) HARLDQ motif mutant S60/R118H/Q121H/K254H | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
4Q33
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110 | | Descriptor: | 4-[(1R)-1-[1-(4-chlorophenyl)-1,2,3-triazol-4-yl]ethoxy]-1-oxidanyl-quinoline, ACETIC ACID, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110
TO BE PUBLISHED
|
|
5LYR
 
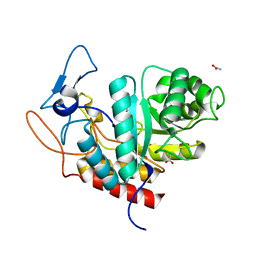 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
3WRT
 
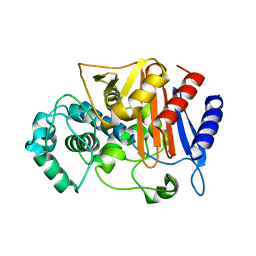 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5IYV
 
 | |
5M42
 
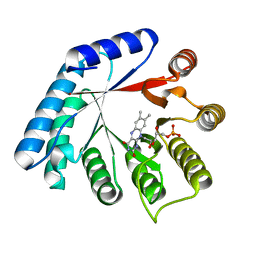 | | Structure of Thermus thermophilus L-proline dehydrogenase lacking alpha helices A, B and C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Proline dehydrogenase | | Authors: | Martinez-Julvez, M, Huijbers, M.M.E, van Berkel, W.J.H, Medina, M. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline dehydrogenase from Thermus thermophilus does not discriminate between FAD and FMN as cofactor.
Sci Rep, 7, 2017
|
|
