6YR9
 
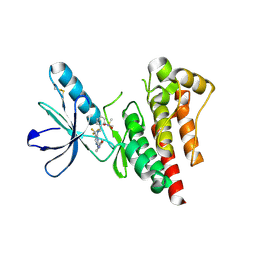 | |
2H2I
 
 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
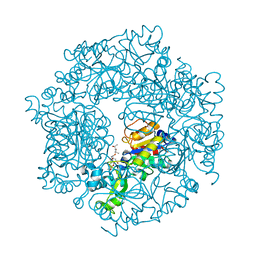
6Q2W
 
 | | Crystal structure of human ROR gamma LBD in complex with a quinoline sulfonamide inverse agonist | | Descriptor: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[4-imidazol-1-yl-2-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]sulfonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Ciesielski, F, Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of novel quinoline sulphonamide derivatives as potent, selective and orally active ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
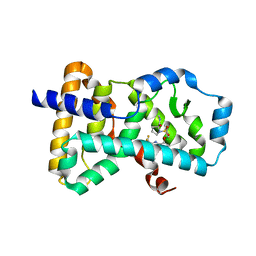
6NB3
 
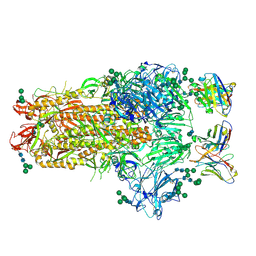 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
2PD0
 
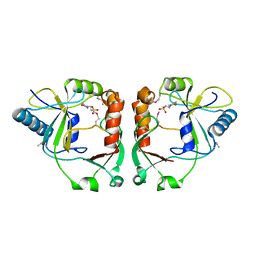 | | Protein cgd2_2020 from Cryptosporidium parvum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein | | Authors: | Cymborowski, M, Chruszcz, M, Hills, T, Lew, J, Melone, M, Zhao, Y, Artz, J, Wernimont, A, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Arrowsmith, C, Hui, R, Minor, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein cgd2_2020 from Cryptosporidium parvum
To be Published
|
|
6BQX
 
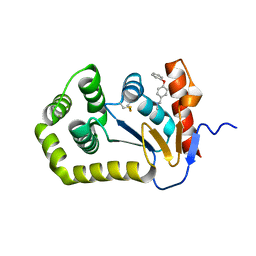 | | Crystal structure of Escherichia coli DsbA in complex with N-methyl-1-(4-phenoxyphenyl)methanamine | | Descriptor: | N-methyl-1-(4-phenoxyphenyl)methanamine, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
5WR9
 
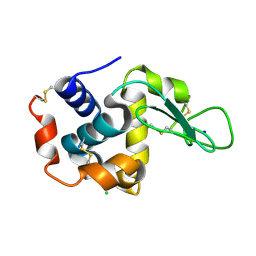 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5N18
 
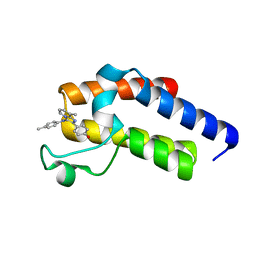 | | Second Bromodomain (BD2) from Candida albicans Bdf1 bound to an imidazopyridine (compound 2) | | Descriptor: | 4-[8-methyl-3-[(4-methylphenyl)amino]imidazo[1,2-a]pyridin-2-yl]phenol, Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
8XB7
 
 | |
5D9B
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
6N89
 
 | |
7BS2
 
 | | Bovine Pancreatic Trypsin with serotonin (Room Temperature) | | Descriptor: | CALCIUM ION, Cationic trypsin, SEROTONIN, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
6MZ3
 
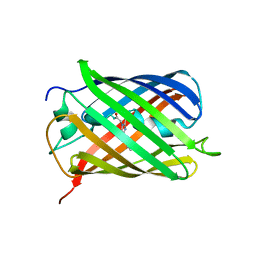 | | mCherry pH sensitive mutant - M66T (mCherryTYG) | | Descriptor: | PAmCherry1 protein | | Authors: | Haynes, E.P, Tantama, M. | | Deposit date: | 2018-11-03 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.088 Å) | | Cite: | Quantifying Acute Fuel and Respiration Dependent pH Homeostasis in Live Cells Using the mCherryTYG Mutant as a Fluorescence Lifetime Sensor.
Anal.Chem., 91, 2019
|
|
6QJ2
 
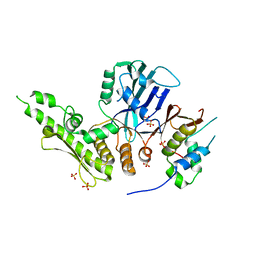 | |
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
8PXV
 
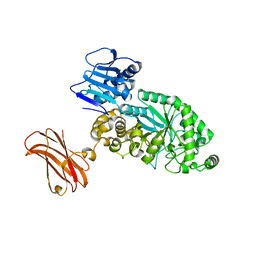 | | Targeting extended blood antigens by Akkermansia muciniphila enzymes unveils a missing link for generating universal donor blood | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL, SODIUM ION, ... | | Authors: | Weikum, J, Jensen, M, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Akkermansia muciniphila exoglycosidases target extended blood group antigens to generate ABO-universal blood.
Nat Microbiol, 9, 2024
|
|
6YJI
 
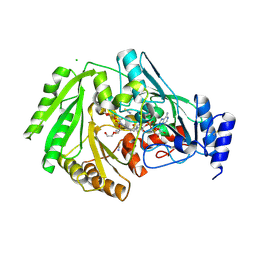 | | Structure of FgCelDH7C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Haddad Momeni, M, Fredslund, F, Berrin, J.G, Abou Hachem, M, Welner, D.H. | | Deposit date: | 2020-04-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs.
Nat Commun, 12, 2021
|
|
5FT6
 
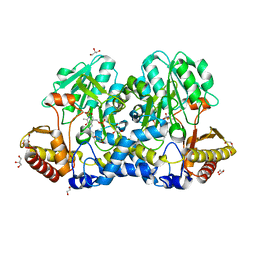 | | Crystal structure of the cysteine desulfurase CsdA (S-sulfonic acid) from Escherichia coli at 2.050 Angstroem resolution | | Descriptor: | CYSTEINE DESULFURASE CSDA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
6YJO
 
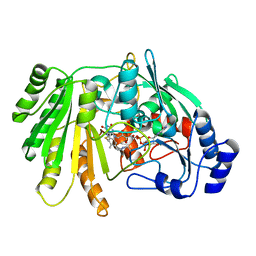 | | Structure of FgChi7B | | Descriptor: | FAD-binding PCMH-type domain-containing protein, FLAVIN-ADENINE DINUCLEOTIDE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Haddad Momeni, M, Fredslund, F, Berrin, J.G, Abou Hachem, M, Welner, D.H. | | Deposit date: | 2020-04-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs.
Nat Commun, 12, 2021
|
|
5JA7
 
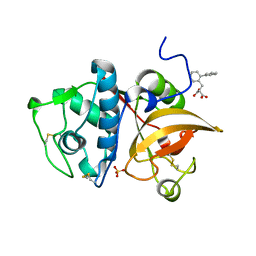 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC94914 | | Descriptor: | ACETATE ION, Cathepsin K, GLYCEROL, ... | | Authors: | Novinec, M, Korenc, M, Lenarcic, B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An allosteric site enables fine-tuning of cathepsin K by diverse effectors.
FEBS Lett., 590, 2016
|
|
5N91
 
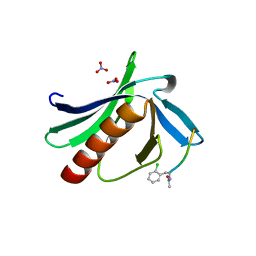 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPP-OH | | Descriptor: | Ac-[2-Cl-F]-PPPP-OH, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Q7L
 
 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
8K58
 
 | | The cryo-EM map of close TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
7BS1
 
 | | Bovine Pancreatic Trypsin with benzamidine (Room Temperature) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
