2WPE
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073359) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
3S2F
 
 | | Crystal Structure of FurX NADH:Furfural | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHORYLISOPROPANE, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
4X05
 
 | |
1WWL
 
 | | Crystal structure of CD14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monocyte differentiation antigen CD14 | | Authors: | Kim, J.-I, Lee, C.J, Jin, M.S, Lee, C.-H, Paik, S.-G, Lee, H, Lee, J.-O. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling
J.Biol.Chem., 280, 2005
|
|
8AO1
 
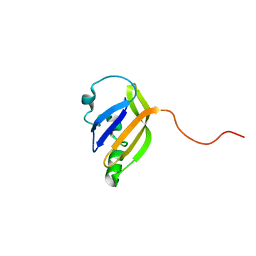 | | solution structure of nanoFAST fluorogen-activating protein in the apo state | | Descriptor: | nanoFAST | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
8CRL
 
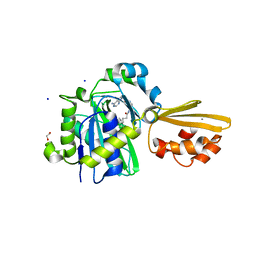 | | Crystal structure of LplA1 in complex with the inhibitor C3 (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8AO0
 
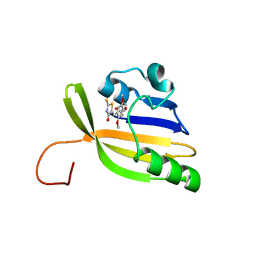 | | Solution structure of nanoFAST/HBR-DOM2 complex | | Descriptor: | (5~{Z})-5-[(2,5-dimethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Photoactive yellow protein | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baleeva, N.S, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
8CRI
 
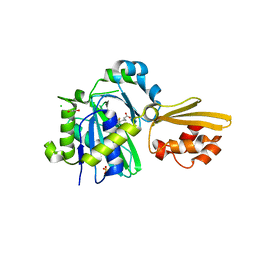 | | Crystal structure of LplA1 in complex with lipoic acid (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LIPOIC ACID, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRJ
 
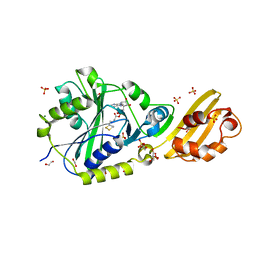 | | Crystal structure of LplA1 in complex with lipoyl-AMP (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6MST
 
 | | Cryo-EM structure of human AA amyloid fibril | | Descriptor: | Serum amyloid A-1 protein | | Authors: | Loerch, S, Rennegarbe, M, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
1XD8
 
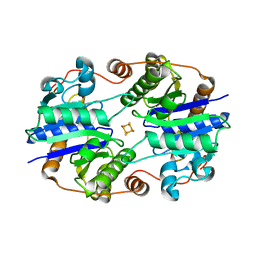 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
8FYG
 
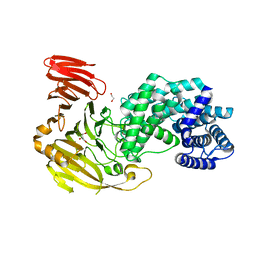 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
3S2E
 
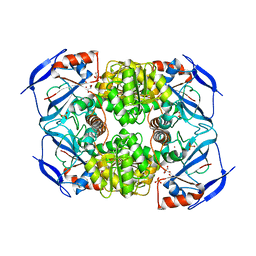 | | Crystal Structure of FurX NADH Complex 1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal Structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
1XA1
 
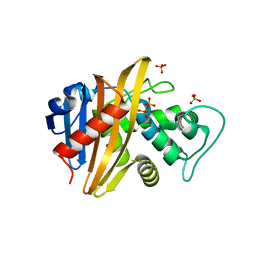 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
8FNX
 
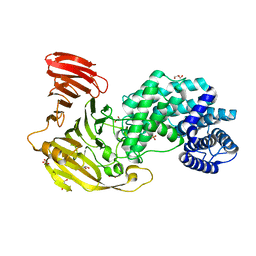 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
3SIH
 
 | |
4XJX
 
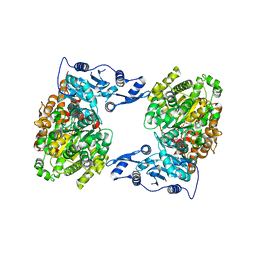 | | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HsdR, MAGNESIUM ION | | Authors: | Baikova, T, Stsiapanava, A, Moche, M, Degtjarik, O, Kuta-Smatanova, I, Ettrich, R. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP
To Be Published
|
|
1XCP
 
 | |
8AGG
 
 | |
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1X78
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1, [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
8ALK
 
 | | Structure of the Legionella phosphocholine hydrolase Lem3 in complex with its substrate Rab1 | | Descriptor: | 2-[[[5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxyethyl-[3-(2-chloranylethanoylamino)propyl]-dimethyl-azanium, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kaspers, M.S, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
1XE0
 
 | | The structure and function of Xenopus NO38-core, a histone binding chaperone in the nucleolus | | Descriptor: | Nucleophosmin | | Authors: | Namboodiri, V.M, Akey, I.V, Schmidt-Zachmann, M.S, Head, J.F, Akey, C.W. | | Deposit date: | 2004-09-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure and Function of Xenopus NO38-Core, a Histone Chaperone in the Nucleolus.
Structure, 12, 2004
|
|
8CRG
 
 | | E. coli adenylate kinase in complex with two ADP molecules as a result of enzymatic AP4A hydrolysis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase | | Authors: | Oelker, M, Tischlik, S, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Insights into Enzymatic Catalysis from Binding and Hydrolysis of Diadenosine Tetraphosphate by E. coli Adenylate Kinase.
Biochemistry, 62, 2023
|
|
6QEG
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 2-Oxo-1-phenyl-pyrrolidine-3-carboxylic acid (2-thiophen-2-yl-ethyl)-amide | | Descriptor: | (3~{S})-3-oxidanyl-2-oxidanylidene-1-phenyl-~{N}-(2-thiophen-2-ylethyl)pyrrolidine-3-carboxamide, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
