3D37
 
 | | The crystal structure of the tail protein from Neisseria meningitidis MC58 | | Descriptor: | CHLORIDE ION, Tail protein, 43 kDa | | Authors: | Zhang, R, Li, H, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the tail protein from Neisseria meningitidis MC58.
To be Published
|
|
7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | Descriptor: | L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7Z3J
 
 | | Structure of crystallisable rat Phospholipase C gamma 1 in complex with inositol 1,4,5-trisphosphate | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Pinotsis, N, Bunney, T.D, Katan, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the membrane interactions of phospholipase C gamma reveals key features of the active enzyme.
Sci Adv, 8, 2022
|
|
3E5O
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7F1U
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-methionine intermediates | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, L-methionine gamma-lyase, METHIONINE | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
3E8P
 
 | | Crystal structure of the protein Q8E9M7 from Shewanella oneidensis related to thioesterase superfamily. Northeast Structural Genomics Consortium target SoR246. | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Janjua, H, Ciccosanti, C, Foote, E.L, Wang, H, Xiao, R, Nair, R, Everett, J, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the protein Q8E9M7 from Shewanella oneidensis related to thioesterase superfamily. Northeast Structural Genomics Consortium target SoR246.
To be Published
|
|
3EFB
 
 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | Descriptor: | ACETIC ACID, Probable sor-operon regulator | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
7F1V
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-homocysteine intermediates | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
3EID
 
 | | CDK2/CyclinA complexed with a pyrazolopyridazine inhibitor | | Descriptor: | (2S)-1-(dimethylamino)-3-(4-{[4-(6-morpholin-4-ylpyrazolo[1,5-b]pyridazin-3-yl)pyrimidin-2-yl]amino}phenoxy)propan-2-ol, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Steven, K, Reno, M, Alberti, J, Price, D, Kane-Carson, L, Knick, V, Shewchuk, L, Hassell, A, Veal, J, Peel, M. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and evaluation of pyrazolo[1,5-b]pyridazines as selective cyclin dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7ZCE
 
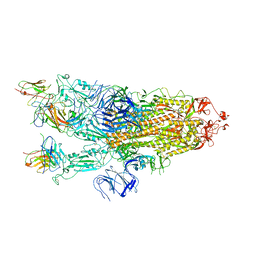 | |
3EIV
 
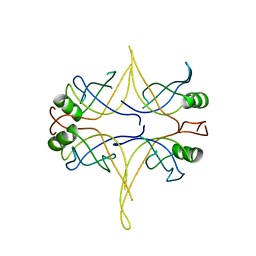 | |
7FEV
 
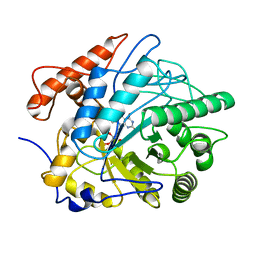 | | Crystal structure of Old Yellow Enzyme6 (OYE6) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN binding | | Authors: | Singh, Y, Sharma, R, Mishra, M, Verma, P.K, Saxena, A.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Crystal structure of ArOYE6 reveals a novel C-terminal helical extension and mechanistic insights into the distinct class III OYEs from pathogenic fungi.
Febs J., 289, 2022
|
|
7EXK
 
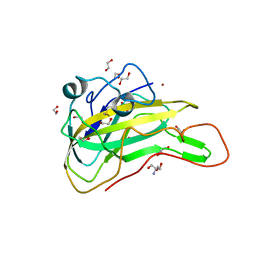 | | An AA9 LPMO of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nguyen, H, Kondo, K, Nagata, T, Katahira, M, Mikami, B. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional and Structural Characterizations of Lytic Polysaccharide Monooxygenase, Which Cooperates Synergistically with Cellulases, from Ceriporiopsis subvermispora.
Acs Sustain Chem Eng, 10, 2022
|
|
3EOK
 
 | |
7Z39
 
 | | Structure of Belumosudil bound to CK2alpha | | Descriptor: | 2-[3-[4-(1~{H}-indazol-5-ylamino)quinazolin-2-yl]phenoxy]-~{N}-propan-2-yl-ethanamide, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the Rho-associated coiled-coil kinase 2 inhibitor belumosudil bound to CK2 alpha.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ZCF
 
 | |
3ERJ
 
 | | Crystal structure of the peptidyl-tRNA hydrolase AF2095 from Archaeglobus fulgidis. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Conover, K, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Cooper, B, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-02 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the peptidyl-tRNA hydrolase AF2095 from Archaeglobus fulgidis. Northeast Structural Genomics Consortium target GR4
To be Published
|
|
3E4N
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 40 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7F5Y
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | FORMIC ACID, Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
7F5Z
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
3E5I
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7Z20
 
 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7Z6N
 
 | | Crystal structure of Zn2+-transporter BbZIP in a metal-stripped state | | Descriptor: | Putative membrane protein, SULFATE ION | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
3E9N
 
 | | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum | | Descriptor: | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum
To be Published
|
|
7Z6M
 
 | | Crystal structure of Zn2+-transporter BbZIP in a cadmium bound state | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
