6MGB
 
 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,4 transferase | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsular polysaccharide export system protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
7C9R
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF THIORHODOVIBRIO STRAIN 970 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26-undecaene, Alpha subunit 1 of light-harvesting 1 complex, ... | | Authors: | Tani, K, Kanno, R, Makino, Y, Hall, M, Takenouchi, M, Imanishi, M, Yu, L.-J, Overmann, J, Madigan, M.T, Kimura, Y, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2020-06-07 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of a Ca 2+ -bound photosynthetic LH1-RC complex containing multiple alpha beta-polypeptides.
Nat Commun, 11, 2020
|
|
4WVB
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Uncharacterized protein, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
4OVU
 
 | | Crystal Structure of p110alpha in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
5YEF
 
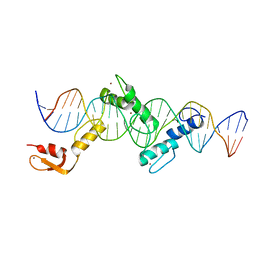 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | Descriptor: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
4X09
 
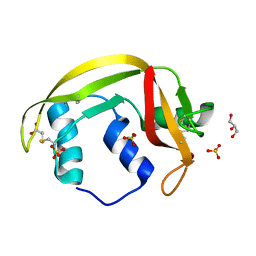 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
4X0J
 
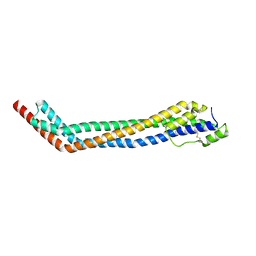 | | Trypanosoma brucei haptoglobin-haemoglobin receptor | | Descriptor: | Haptoglobin-hemoglobin receptor | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
4X52
 
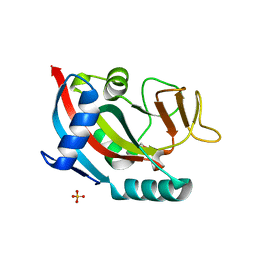 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
4WRI
 
 | | Crystal structure of okadaic acid binding protein 2.1 | | Descriptor: | OKADAIC ACID, Okadaic acid binding protein 2-alpha | | Authors: | Ehara, H, Makino, M, Kodama, K, Ito, T, Sekine, S, Fukuzawa, S, Yokoyama, S, Tachibana, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Okadaic Acid Binding Protein 2.1: A Sponge Protein Implicated in Cytotoxin Accumulation
Chembiochem, 16, 2015
|
|
4WJ4
 
 | | Crystal structure of non-discriminating aspartyl-tRNA synthetase from Pseudomonas aeruginosa complexed with tRNA(Asn) and aspartic acid | | Descriptor: | 76mer-tRNA, ASPARTIC ACID, Aspartate--tRNA(Asp/Asn) ligase | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WJI
 
 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
4WSZ
 
 | |
4WT0
 
 | |
4WT3
 
 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WM9
 
 | | Acinetobacter baumanii OXA-24 complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Mangani, S, Benvenuti, M, Docquier, J.-D. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Selective Inhibition and Slow Reversibility of Avibactam against Class D Carbapenemases: A Structure-Guided Study of OXA-24 and OXA-48.
Acs Chem.Biol., 10, 2015
|
|
4WT4
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT5
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form II | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WNJ
 
 | | Crystal structure of Transthyretin-quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Berni, R, Folli, C. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8HLE
 
 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
4X71
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Shahar, A, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WSD
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the second position in the A-site and with antibiotic paromomycin. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WVZ
 
 | | Crystal structure of artificial crosslinked thiol dioxygenase G95C variant from Pseudomonas aeruginosa | | Descriptor: | 3-mercaptopropionate dioxygenase, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2014-11-09 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Substrate and pH-Dependent Kinetic Profile of 3-Mercaptopropionate Dioxygenase from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
