1MQ5
 
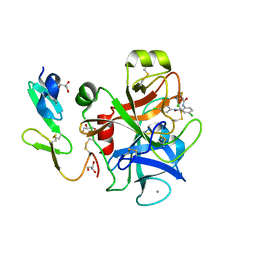 | |
1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
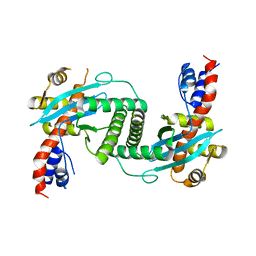
1UOU
 
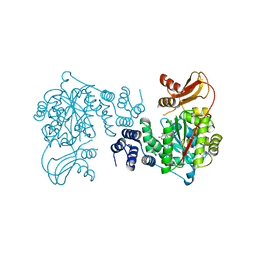 | | Crystal structure of human thymidine phosphorylase in complex with a small molecule inhibitor | | Descriptor: | 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, THYMIDINE PHOSPHORYLASE | | Authors: | Norman, R.A, Barry, S.T, Bate, M, Breed, J, Colls, J.G, Ernill, R.J, Luke, R.W.A, Minshull, C.A, McAlister, M.S.B, McCall, E.J, McMiken, H.H.J, Paterson, D.S, Timms, D, Tucker, J.A, Pauptit, R.A. | | Deposit date: | 2003-09-23 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of Human Thymidine Phosphorylase in Complex with a Small Molecule Inhibitor
Structure, 12, 2004
|
|
3KVW
 
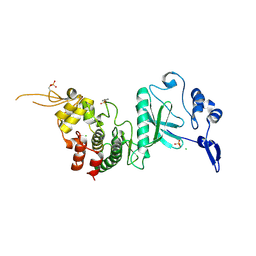 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
4OS3
 
 | | Three-dimensional structure of the C65A-W112F double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7MU8
 
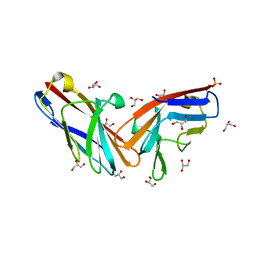 | | Structure of the minimally glycosylated human CEACAM1 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, ... | | Authors: | Belcher Dufrisne, M, Swope, N, Kieber, M, Yang, J.Y, Han, J, Li, J, Moremen, K.W, Prestegard, J.H, Columbus, L. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human CEACAM1 N-domain dimerization is independent from glycan modifications.
Structure, 30, 2022
|
|
5NM2
 
 | | A2A Adenosine receptor cryo structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5ULL
 
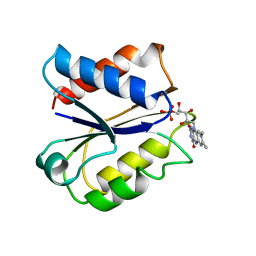 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: REDUCED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
3L19
 
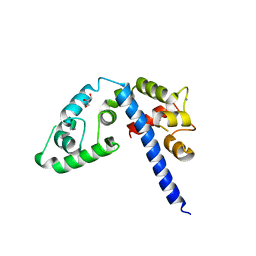 | | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820 | | Descriptor: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, GLYCEROL, ... | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820
To be Published
|
|
1VSR
 
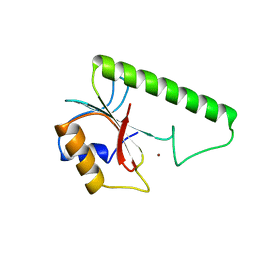 | | VERY SHORT PATCH REPAIR (VSR) ENDONUCLEASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (VSR ENDONUCLEASE), ZINC ION | | Authors: | Tsutakawa, S.E, Muto, T, Jingami, H, Kunishima, N, Ariyoshi, M, Kohda, D, Nakagawa, M, Morikawa, K. | | Deposit date: | 1999-02-13 | | Release date: | 1999-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and functional studies of very short patch repair endonuclease.
Mol.Cell, 3, 1999
|
|
4ORW
 
 | | Three-dimensional structure of the C65A-K59A double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OS8
 
 | | Three-dimensional structure of the C65A-W54F-W112F triple mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1W24
 
 | | Crystal Structure Of human Vps29 | | Descriptor: | VACUOLAR PROTEIN SORTING PROTEIN 29 | | Authors: | Wang, D, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2004-06-26 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Vacuolar Protein Sorting Protein 29 Reveals a Phosphodiesterase/Nuclease-Like Fold and Two Protein-Protein Interaction Sites.
J.Biol.Chem., 280, 2005
|
|
3C0V
 
 | | Crystal structure of cytokinin-specific binding protein in complex with cytokinin and Ta6Br12 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytokinin-specific binding protein, ... | | Authors: | Pasternak, O, Bujacz, A, Biesiadka, J, Bujacz, G, Sikorski, M, Jaskolski, M. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MAD phasing using the (Ta(6)Br(12))(2+) cluster: a retrospective study
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5GV0
 
 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
1VEF
 
 | | Acetylornithine aminotransferase from Thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Three-Dimensional Strutcure of Acetylornithine aminotransferase from Thermus thermophilus HB8
To be Published
|
|
1VGI
 
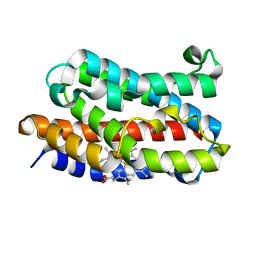 | | Crystal structure of xenon bound rat heme-heme oxygenase-1 complex | | Descriptor: | FORMIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
3KRR
 
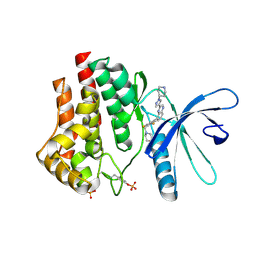 | | Crystal Structure of JAK2 complexed with a potent quinoxaline ATP site inhibitor | | Descriptor: | 8-[3,5-difluoro-4-(morpholin-4-ylmethyl)phenyl]-2-(1-piperidin-4-yl-1H-pyrazol-4-yl)quinoxaline, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Gerspacher, M, Kroemer, M, Scheufler, C. | | Deposit date: | 2009-11-19 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibition of Polycythemia by the Quinoxaline JAK2 Inhibitor NVP-BSK805
Mol.Cancer Ther., 9, 2010
|
|
5NP6
 
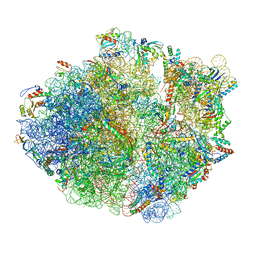 | | 70S structure prior to bypassing | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Klimova, M, Zamora, M, Gil-Carton, D, Rodnina, M, Valle, M. | | Deposit date: | 2017-04-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome rearrangements at the onset of translational bypassing.
Sci Adv, 3, 2017
|
|
3CB5
 
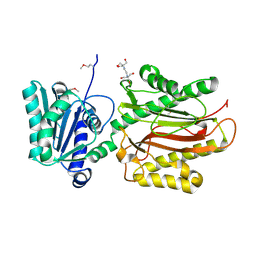 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form A) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5N8V
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1-(2-azanylethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
6X2K
 
 | |
5N2C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, ... | | Authors: | Matterazzo, E, Bolognesi, M, Gourlay, L.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing Probes for Immunodiagnostics: Structural Insights into an Epitope Targeting Burkholderia Infections.
ACS Infect Dis, 3, 2017
|
|
6WJ5
 
 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
6DSO
 
 | | Cryo-EM structure of murine AA amyloid fibril | | Descriptor: | Serum amyloid A-2 protein | | Authors: | Loerch, S, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-06-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
