5B2J
 
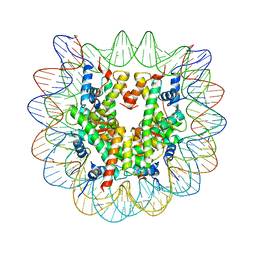 | | Human nucleosome containing CpG methylated DNA | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Fujii, Y, Wakamori, M, Umehara, T, Yokoyama, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human nucleosome core particle containing enzymatically introduced CpG methylation.
Febs Open Bio, 6, 2016
|
|
4WRY
 
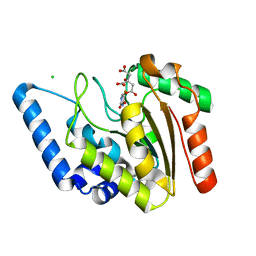 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil(B), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3MNM
 
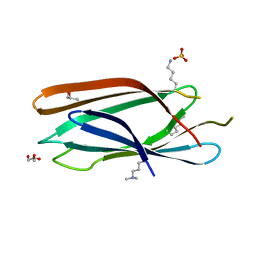 | | Crystal structure of GAE domain of GGA2p from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-binding protein GGA2, GLYCEROL, SULFATE ION | | Authors: | Fang, P, Wang, J, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-08 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for the specificity of the GAE domain of yGGA2 for its accessory proteins Ent3 and Ent5
Biochemistry, 49, 2010
|
|
4N0E
 
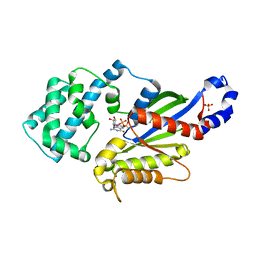 | | Crystal structure of the K345L variant of the Gi alpha1 subunit bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, SULFATE ION | | Authors: | Thaker, T.M, Preininger, A.M, Sarwar, M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2013-10-01 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Transient Interaction between the Phosphate Binding Loop and Switch I Contributes to the Allosteric Network between Receptor and Nucleotide in G alpha i1.
J.Biol.Chem., 289, 2014
|
|
6IBV
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
4WS1
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (AB), Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1KPL
 
 | | Crystal Structure of the ClC Chloride Channel from S. typhimurium | | Descriptor: | CHLORIDE ION, N-OCTANE, PENTADECANE, ... | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
7ZLA
 
 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
6GBQ
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Reston virus | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
1KH2
 
 | |
4WS5
 
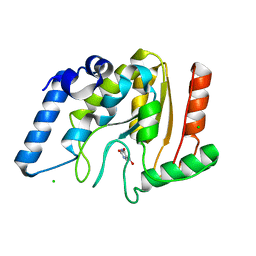 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-nitrouracil, Form III | | Descriptor: | 5-nitrouracil, CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4N0M
 
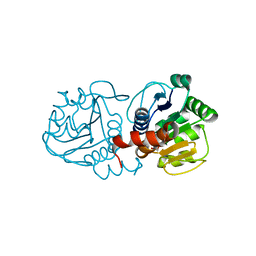 | | Crystal structure of human C53A DJ-1 in complex with Cu | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-10-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
1KQF
 
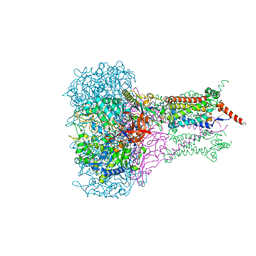 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARDIOLIPIN, FORMATE DEHYDROGENASE, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
7ZPA
 
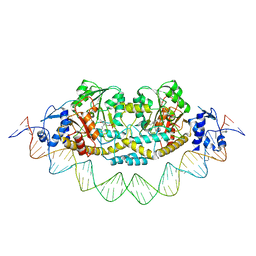 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
2BYD
 
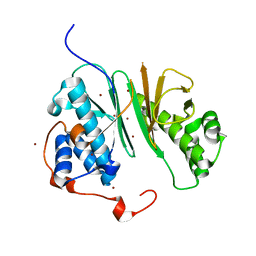 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase | | Descriptor: | BROMIDE ION, HSPC223 | | Authors: | Bunkoczi, G, Wu, X, Dubinina, E, Johansson, C, Smee, C, Turnbull, A, Oppermann, U, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-29 | | Release date: | 2005-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and substrate recognition of human holo ACP synthase.
Chem. Biol., 14, 2007
|
|
1KSM
 
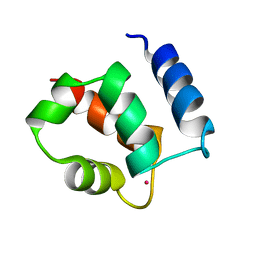 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | Deposit date: | 2002-01-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
3M7I
 
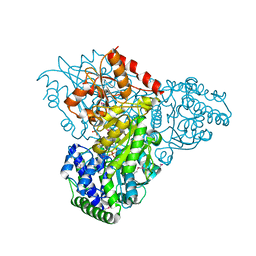 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 5-O-phosphono-beta-D-ribofuranose, MAGNESIUM ION, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion
TO BE PUBLISHED
|
|
4WUA
 
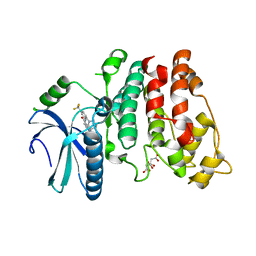 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | Descriptor: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | Authors: | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
3LZN
 
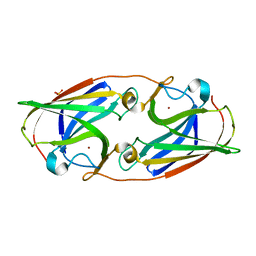 | | Crystal Structure Analysis of the apo P19 protein from Campylobacter jejuni at 1.59 A at pH 9 | | Descriptor: | P19 protein, SULFATE ION, ZINC ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
6FXY
 
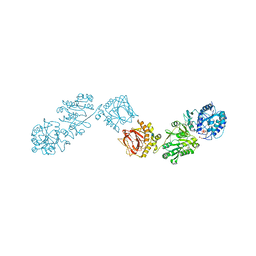 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal - Structure from long-wavelength S-SAD | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
3LZV
 
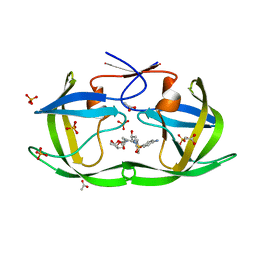 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Kolli, M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
6IN0
 
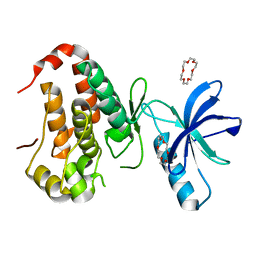 | | Crystal structure of EphA3 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Ephrin type-A receptor 3 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6FZ1
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
1KWK
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
