5O2Z
 
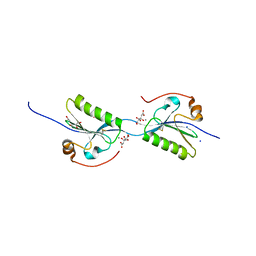 | | Domain swap dimer of the G167R variant of gelsolin second domain | | Descriptor: | ACETATE ION, CALCIUM ION, CITRATE ANION, ... | | Authors: | Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein.
Hum. Mol. Genet., 27, 2018
|
|
6V5M
 
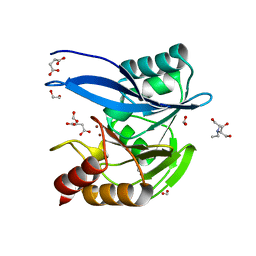 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
1Y2V
 
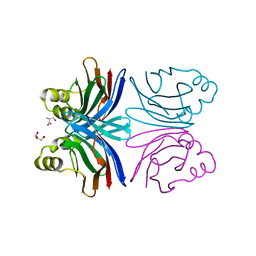 | | Crystal structure of the common edible mushroom (Agaricus bisporus) lectin in complex with T-antigen | | Descriptor: | SERINE, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin | | Authors: | Carrizo, M.E, Capaldi, S, Perduca, M, Irazoqui, F.J, Nores, G.A, Monaco, H.L. | | Deposit date: | 2004-11-23 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Antineoplastic Lectin of the Common Edible Mushroom (Agaricus bisporus) Has Two Binding Sites, Each Specific for a Different Configuration at a Single Epimeric Hydroxyl
J.Biol.Chem., 280, 2005
|
|
5H4Z
 
 | | Crystal structure of S202G mutant of human SYT-5 C2A domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Synaptotagmin-5 | | Authors: | Qiu, X, Ge, J, Yan, X, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2016-11-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural analysis of Ca(2+)-binding pocket of synaptotagmin 5 C2A domain
Int. J. Biol. Macromol., 95, 2017
|
|
3ZX0
 
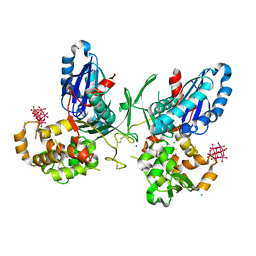 | | NTPDase1 in complex with Heptamolybdate | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for a domain motion in rat nucleoside triphosphate diphosphohydrolase (NTPDase) 1.
J. Mol. Biol., 415, 2012
|
|
7QHY
 
 | | Structure of a Kluyveromyces lactis protein involved in RNA decay | | Descriptor: | GLYCEROL, Nonsense-mediated decay protein 4,Nonsense-mediated decay protein 4,Nonsense mediated mRNA decay protein 4 (Nmd4), SULFATE ION | | Authors: | Barbarin-Bocahu, I, Graille, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The X-ray crystallography phase problem solved thanks to AlphaFold and RoseTTAFold models: a case-study report.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6VHF
 
 | | Crystal structure of RbBP5 interacting domain of Cfp1 | | Descriptor: | PHD-type domain-containing protein, ZINC ION | | Authors: | Joshi, M, Couture, J.F. | | Deposit date: | 2020-01-09 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | A non-canonical monovalent zinc finger stabilizes the integration of Cfp1 into the H3K4 methyltransferase complex COMPASS.
Nucleic Acids Res., 48, 2020
|
|
7Q82
 
 | |
6BZW
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody AP33 | | Descriptor: | AP33 GL Heavy Chain, AP33 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7Q7Y
 
 | |
5GSM
 
 | | Glycoside hydrolase B with product | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Watanabe, M, Kamachi, S, Mine, S. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Glycoside hydrolase B with product
To Be Published
|
|
8BN8
 
 | | METTL3-METTL14 heterodimer bound to the SAM competitive small molecule inhibitor STM3006 | | Descriptor: | 2-[[4-(6-bromanyl-2~{H}-indazol-4-yl)-1,2,3-triazol-1-yl]methyl]-6-[(4,4-dimethylpiperidin-1-yl)methyl]imidazo[1,2-a]pyridine, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit | | Authors: | Pilka, E.S, Thomas, B, Blackaby, W, Hardick, D, Feeney, K, Ridgill, M, Rotty, B, Rausch, O. | | Deposit date: | 2022-11-13 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Inhibition of METTL3 Results in a Cell-Intrinsic Interferon Response That Enhances Antitumor Immunity.
Cancer Discov, 13, 2023
|
|
1VTE
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
7QDW
 
 | |
6C1T
 
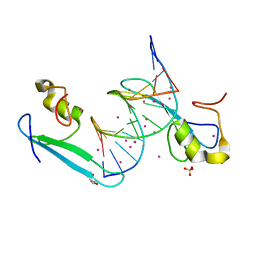 | | MBD2 in complex with a partially methylated DNA | | Descriptor: | 12-mer DNA, GLYCEROL, Methyl-CpG-binding domain protein 2, ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
3ZTR
 
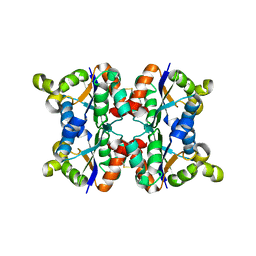 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FIRST STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
6C24
 
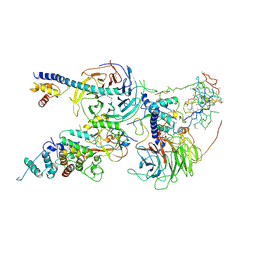 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
5GUE
 
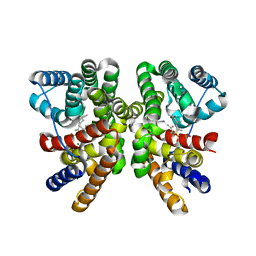 | | Crystal structure of CotB2 (GGSPP/Mg2+-Bound Form) from Streptomyces melanosporofaciens | | Descriptor: | Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION, phosphonooxy-[(10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenyl]sulfanyl-phosphinic acid | | Authors: | Tomita, T, Kim, S.-Y, Ozaki, T, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2016-08-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the CotB2-Catalyzed Cyclization of Geranylgeranyl Diphosphate to the Diterpene Cyclooctat-9-en-7-ol
ACS Chem. Biol., 12, 2017
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
7QTO
 
 | |
5NUB
 
 | |
3MQM
 
 | | Crystal Structure of the Bromodomain of human ASH1L | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Felletar, I, Vollmar, M, Chaikuad, A, Krojer, T, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7Q2A
 
 | | Crystal structure of AphC in complex with 4-ethylcatechol | | Descriptor: | 4-ethylbenzene-1,2-diol, CALCIUM ION, Catechol 2,3-dioxygenase, ... | | Authors: | Zahn, M, Grigg, J.C, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of a phylogenetically distinct extradiol dioxygenase involved in the bacterial catabolism of lignin-derived aromatic compounds.
J.Biol.Chem., 298, 2022
|
|
7QBK
 
 | | Crystal structure of a second homolog of R2-like ligand-binding oxidase in Sulfolobus acidocaldarius (SaR2loxII) | | Descriptor: | FE (III) ION, MANGANESE (III) ION, R2-like ligand-binding oxidase (homolog II) from Sulfolobus acidocaldarius | | Authors: | Lebrette, H, Diamanti, R, Srinivas, V, Hogbom, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Febs Lett., 596, 2022
|
|
7QTU
 
 | |
