2YQ5
 
 | | Crystal Structure of D-isomer specific 2-hydroxyacid dehydrogenase from Lactobacillus delbrueckii ssp. bulgaricus: NAD complexed form | | Descriptor: | D-ISOMER SPECIFIC 2-HYDROXYACID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Holton, S.J, Anandhakrishnan, M, Geerlof, A, Wilmanns, M. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Characterization of D-Isomer Specific 2-Hydroxyacid Dehydrogenase from Lactobacillus Delbrueckii Ssp. Bulgaricus
J.Struct.Biol., 181, 2013
|
|
5F0J
 
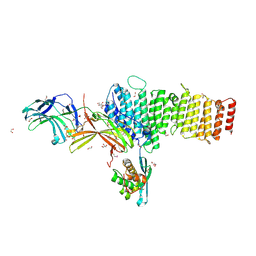 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5J5H
 
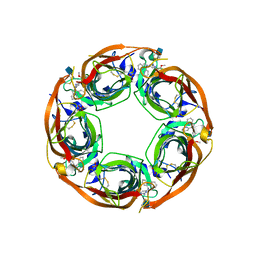 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 6-(2-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(2-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6YNC
 
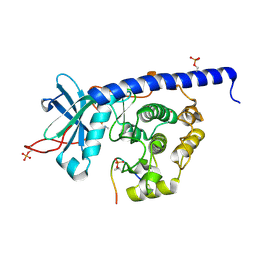 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with the methylated Fasudil-derived fragment N-methylisoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-methylisoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
5ET1
 
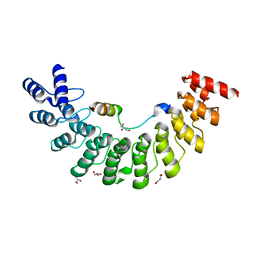 | | Crystal structure of Myo3b-ARB1 in complex with Espin1-AR | | Descriptor: | Espin, GLYCEROL, Myosin-IIIb | | Authors: | Liu, H, Li, J, liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
6YQ2
 
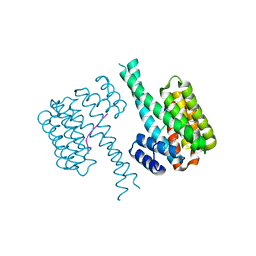 | |
2YNW
 
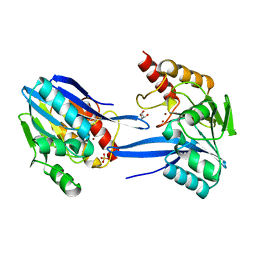 | | GIM-1-2Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, SULFATE ION, ... | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
5J6P
 
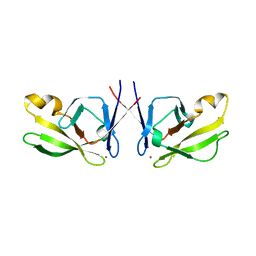 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | Descriptor: | Kinetochore protein mis18, ZINC ION | | Authors: | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | Deposit date: | 2016-04-05 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
4GY9
 
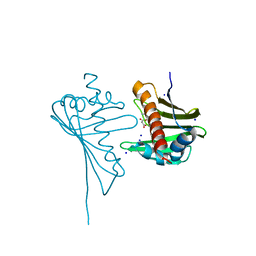 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6YQI
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with long-chain Fasudil-derivative N-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
2YOZ
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, crystallized with 2'-AMPS | | Descriptor: | 2'-O-(sulfidophosphinato)adenosine, 2,3-CYCLIC NUCLEOTIDE 3'-PHOSPHODIESTERASE, ACETATE ION, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-10-29 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
1JS0
 
 | | Crystal Structure of 3D Domain-swapped RNase A Minor Trimer | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Gotte, G, Libonati, M, Eisenberg, D. | | Deposit date: | 2001-08-15 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the two 3D domain-swapped RNase A trimers.
Protein Sci., 11, 2002
|
|
3FMN
 
 | | P38 kinase crystal structure in complex with RO2530 | | Descriptor: | 6-(2,4-difluorophenoxy)-N-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-amine, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Ghate, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P38 kinase crystal structure in complex with RO2530
To be Published
|
|
2YPC
 
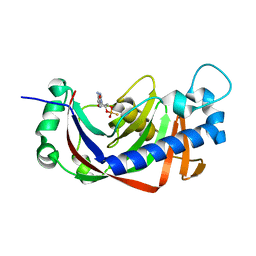 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H309S, crystallized with 2',3-(SP)-Cyclic-AMPS | | Descriptor: | 2', 3'-CYCLIC NUCLEOTIDE 3'-PHOSPHODIESTERASE, [(3aR,4R,6R,6aR)-4-(6-aminopurin-9-yl)-2-oxidanyl-2-sulfanylidene-3a,4,6,6a-tetrahydrofuro[3,4-d][1,3,2]dioxaphosphol-6-yl]methanol | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
6YPG
 
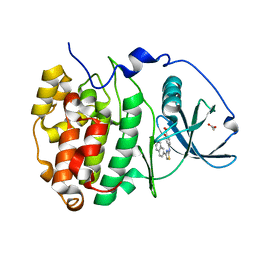 | |
6YPK
 
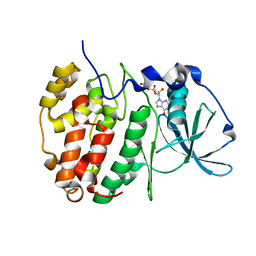 | | Crystal Structure of CK2alpha with GTP bound | | Descriptor: | Casein kinase II subunit alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
5ES4
 
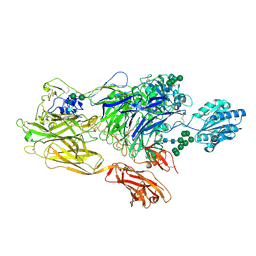 | | RE-REFINEMENT OF INTEGRIN ALPHAXBETA2 ECTODOMAIN IN THE CLOSED/BENT CONFORMATION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sen, M, Springer, T.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-03-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Leukocyte integrin alpha L beta 2 headpiece structures: The alpha I domain, the pocket for the internal ligand, and concerted movements of its loops.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2Y6H
 
 | | X-2 L110F CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
5EVL
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum | | Descriptor: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum
To Be Published
|
|
8OFI
 
 | | Ivabradine bound to HCN4 channel | | Descriptor: | Ivabradine, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Saponaro, A, Chaves-Sanjuan, A, Sharifzadeh, A.S, Clarke, O.B, Marabelli, C, Bolognesi, M, Thiel, G, Moroni, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural determinants of ivabradine block of the open pore of HCN4.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5EX7
 
 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
8K0F
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
6YFG
 
 | | Virus-like particle of Beihai levi-like virus 32 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.897 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
2YND
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyrazole sulphonamide inhibitor. | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
