5J60
 
 | | Structure of a thioredoxin reductase from Gloeobacter violaceus | | Descriptor: | CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, TETRAETHYLENE GLYCOL, ... | | Authors: | Buey, R.M, de Pereda, J.M, Balsera, M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Member of the Thioredoxin Reductase Family from Early Oxygenic Photosynthetic Organisms.
Mol Plant, 10, 2017
|
|
7MS6
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS7
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MUA
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7NK3
 
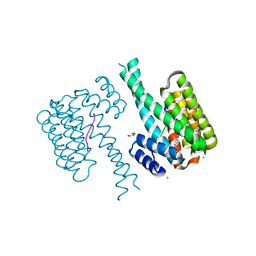 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-128 | | Descriptor: | 14-3-3 protein sigma, 2-(4-methylphenyl)sulfonyl-3,4-dihydro-1~{H}-isoquinoline, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NIF
 
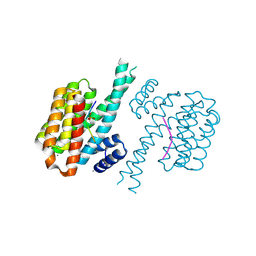 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-011 | | Descriptor: | 1-(4-methylphenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7VK2
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.0 A resolution -9 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Gul, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VB6
 
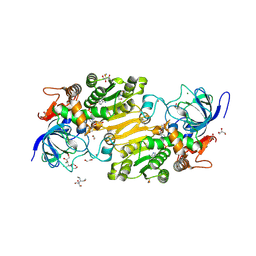 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | Descriptor: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
7NJ6
 
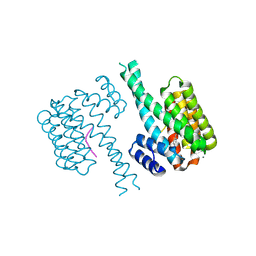 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1005 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(trifluoromethyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NLA
 
 | |
5T8R
 
 | | Crystal structure of human BAZ2A PHD zinc finger in complex with unmodified H3 10-mer | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, Histone H3.1, ... | | Authors: | Amato, A, Gadd, M.S, Bortoluzzi, A, Ciulli, A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of molecular recognition of helical histone H3 tail by PHD finger domains.
Biochem. J., 474, 2017
|
|
5J8J
 
 | | A histone deacetylase from Saccharomyces cerevisiae | | Descriptor: | Histone deacetylase HDA1 | | Authors: | Zhu, Y, Shen, H, Li, X, Teng, M. | | Deposit date: | 2016-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Structural and histone binding ability characterization of the ARB2 domain of a histone deacetylase Hda1 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
7VB3
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aliphatic (R)-hydroxynitrile lyase, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
7NJA
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1006 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5JA0
 
 | | Crystal structure of human FPPS with allosterically bound FPP | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Zielinski, M, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product.
Nat Commun, 8, 2017
|
|
7NH6
 
 | | Crystal structure of human carbonic anhydrase II with 3-(3-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methyl)ureido)benzenesulfonamide | | Descriptor: | 3-(3-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methyl)ureido)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
7VB5
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum complexed with acetone cyanohydrin | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXY-2-METHYLPROPANENITRILE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
7NH8
 
 | | Crystal structure of human carbonic anhydrase II with N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Synthesis of Azasugar-Sulfonamide conjugates and their Evaluation as Inhibitors of Carbonic Anhydrases: the Azasugar Approach to Selectivity
Eur.J.Org.Chem., 2021
|
|
6SP7
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with Taniborbactam (VNRX-5133) | | Descriptor: | (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections.
J.Med.Chem., 63, 2020
|
|
5I36
 
 | | Crystal structure of color device state A | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Kristiansen, M, Sha, R, Birktoft, J, Mao, C, Seeman, N.C. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.123 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7VR9
 
 | | Crystal structure of human serum albumin complex with aripiprazole and myristic acid | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, MYRISTIC ACID, Serum albumin | | Authors: | Kawai, A, Yamasaki, K, Otagiri, M. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of Myristate on the Induced Circular Dichroism Spectra of Aripiprazole Bound to Human Serum Albumin: A Structural-Chemical Investigation
Acs Omega, 7, 2022
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5OCI
 
 | | Human Heat Shock Protein 90 bound to 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid methyl-(4-morpholin-4-yl-phenyl)-amide | | Descriptor: | 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid methyl-(4-morpholin-4-yl-phenyl)-amide, Heat shock protein HSP 90-alpha | | Authors: | Schuetz, D.A, Richter, L, Amaral, M, Grandits, M, Musil, D, Graedler, U, Frech, M, Ecker, G.F. | | Deposit date: | 2017-07-03 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
