5NHT
 
 | | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
6HVG
 
 | |
8GSS
 
 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
4L6E
 
 | | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2. Northeast Structural Genomics Consortium (NESG) Target HR9193b | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Kogan, S, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-12 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2.
To be Published
|
|
5AEF
 
 | | Electron cryo-microscopy of an Abeta(1-42)amyloid fibril | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Schmidt, M, Rohou, A, Lasker, K, Yadav, J.K, Schiene-Fischer, C, Fandrich, M, Grigorieff, N. | | Deposit date: | 2015-08-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Peptide Dimer Structure in an Abeta(1-42) Fibril Visualized with Cryo-Em
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8AKP
 
 | | Crystal structure of the catalytic domain of G7048 from Penicillium sumatraense | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, catalytic domain of G7048 | | Authors: | Troilo, F, Scafati, V, Giovannoni, M, Mattei, B, Benedetti, M, Angelucci, F, Di Matteo, A. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of two 1,3-beta-glucan-modifying enzymes from Penicillium sumatraense reveals new insights into 1,3-beta-glucan metabolism of fungal saprotrophs.
Biotechnol Biofuels Bioprod, 15, 2022
|
|
6SQZ
 
 | | Mouse dCTPase in complex with dCMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
1R3G
 
 | | 1.16A X-ray structure of the synthetic DNA fragment with the incorporated 2'-O-[(2-Guanidinium)ethyl]-5-methyluridine residues | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(GMU)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Prakash, T.P, Puschl, A, Lesnik, E, Tereshko, V, Egli, M, Manoharan, M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 2'-O-[2-(Guanidinium)ethyl]-Modified Oligonucleotides: Stabilizing Effect on Duplex and Triplex Structures.
Org.Lett., 6, 2004
|
|
5AFG
 
 | | Structure of the Stapled Peptide Bound to Mdm2 | | Descriptor: | 1,8-DIETHYL-1,8-DIHYDRODIBENZO[3,4:7,8][1,2,3]TRIAZOLO[4',5':5,6]CYCLOOCTA[1,2-D][1,2,3]TRIAZOLE, E3 UBIQUITIN-PROTEIN LIGASE MDM2, STAPLED PEPTIDE | | Authors: | Lau, Y.H, Wu, Y, Rossmann, M, de Andrade, P, Tan, Y.S, McKenzie, G.J, Venkitaraman, A.R, Hyvonen, M, Spring, D.R. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5NGL
 
 | | The endo-beta1,6-glucanase BT3312 | | Descriptor: | Glucosylceramidase, SODIUM ION, beta-D-glucopyranose-(1-6)-1-DEOXYNOJIRIMYCIN | | Authors: | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
5AG8
 
 | | CRYSTAL STRUCTURE OF A MUTANT (665I6H) OF THE C-TERMINAL DOMAIN OF RGPB | | Descriptor: | GINGIPAIN R2, GLYCEROL, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Outer-Membrane Export Signal of Porphyromonas Gingivalis Type Ix Secretion System (T9Ss) is a Conserved C-Terminal Beta-Sandwich Domain.
Sci.Rep., 6, 2016
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
6SZB
 
 | |
6HA1
 
 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1ZDV
 
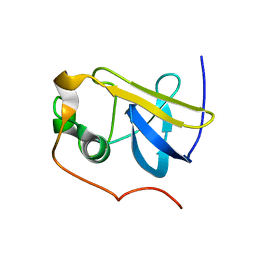 | | Solution Structure of the type 1 pilus assembly platform FimD(25-139) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
5NL8
 
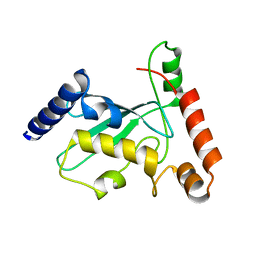 | | Pex4 of Hansenula Polymorpha | | Descriptor: | Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Groves, M, Williams, C. | | Deposit date: | 2017-04-04 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
4ZYB
 
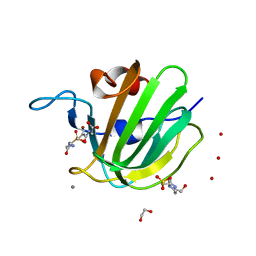 | | High resolution structure of M23 peptidase LytM with substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Grabowska, M, Jagielska, E, Czapinska, H, Bochtler, M, Sabala, I. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of an M23 peptidase with a substrate analogue.
Sci Rep, 5, 2015
|
|
8GI2
 
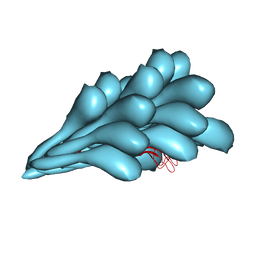 | | Cryo-EM structure of Natrinema sp. J7-2 Type IV pilus | | Descriptor: | Natrinema pilin, Orf10 | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Liu, Y, Krupovic, M, Egelman, E.H. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4ZS9
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sugar binding protein of ABC transporter system, ... | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
8GQ1
 
 | | HyHEL10 Fab complexed with hen egg lysozyme carrying arginine cluster in framework region of light chain. | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C, ... | | Authors: | Matsuura, H, Hirata, K, Sakai, N, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-08-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops.
Protein Sci., 32, 2023
|
|
6XPC
 
 | | Structure of human GGT1 in complex with full GSH molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of glutathione- and inhibitor-bound human GGT1: critical interactions within the cysteinylglycine binding site.
J.Biol.Chem., 296, 2020
|
|
4ZTK
 
 | | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cell division protein FtsI/penicillin binding protein 2 | | Authors: | CUFF, M, OSIPIUK, J, WU, R, ENDRES, M, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila.
to be published
|
|
3N6V
 
 | |
8GRT
 
 | | Small Dipeptide Analogues developed by Co-crystal Structure of Stenotrophomonas maltophilia Dipeptidyl Peptidase 7 | | Descriptor: | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID, Dipeptidyl-peptidase, TYROSINE | | Authors: | Yasumitsu, S, Koushi, H, Akihiro, N, Yoshiyuki, Y, Wataru, O, Mizuki, S, Saori, R, Nobutada, T, Anna, M, Keiko, H, Tsuda, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small Dipeptide Analogues Generated by Co-crystal Structure of Bacterial Dipeptidyl Peptidase 7 to Defeat Stenotrophomonas maltophilia
To Be Published
|
|
1A0B
 
 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB.
Cell(Cambridge,Mass.), 88, 1997
|
|
