4TL0
 
 | |
3P06
 
 | |
4RZD
 
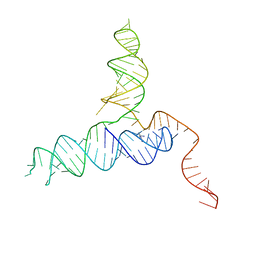 | | Crystal Structure of a PreQ1 Riboswitch | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | Authors: | Wedekind, J.E, Liberman, J.A, Salim, M. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7UDI
 
 | |
4TPH
 
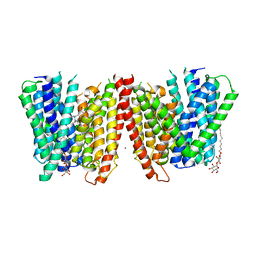 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | Descriptor: | 3,5 DIBROMOTYROSINE, ALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Guettou, F, Quistgaard, E, Raba, M, Moberg, P, Low, C, Nordlund, P. | | Deposit date: | 2014-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6HHN
 
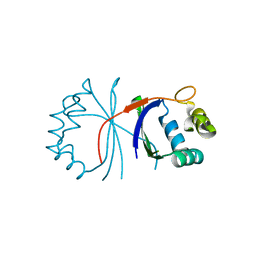 | |
4TQX
 
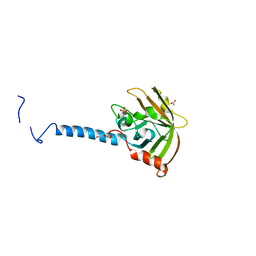 | | Molecular Basis of Streptococcus mutans Sortase A Inhibition by Chalcone. | | Descriptor: | ACETIC ACID, SULFATE ION, Sortase, ... | | Authors: | Wallock-Richards, D.J, Marles-Wright, J, Clarke, D.J, Maitra, A, Dodds, M, Hanley, B, Campopiano, D.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone.
Chem.Commun.(Camb.), 51, 2015
|
|
6HCD
 
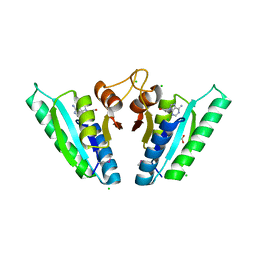 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
4TT6
 
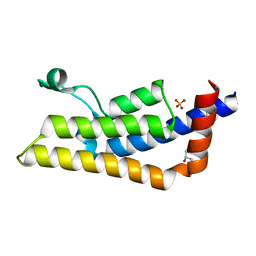 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
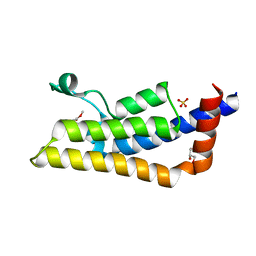 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TOD
 
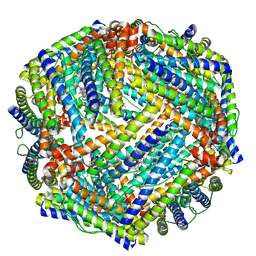 | | 2.05A resolution structure of BfrB (D34F) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TPR
 
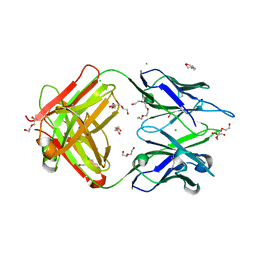 | | Structure of Tau5 antibody Fab fragment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, CHLORIDE ION, ... | | Authors: | Cehlar, O, Skrabana, R, Novak, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Tau5 antibody Fab fragment
To Be Published
|
|
4TSX
 
 | |
6H8T
 
 | | Crystal structure of Papain modify by achiral Ru(II)complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Papain, ... | | Authors: | Cherrier, M.V, Amara, P, Talbi, B, Salmin, M, Fontecilla-Camps, J.C. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic evidence for unexpected selective tyrosine hydroxylations in an aerated achiral Ru-papain conjugate.
Metallomics, 10, 2018
|
|
4RTA
 
 | |
4RNY
 
 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8RBX
 
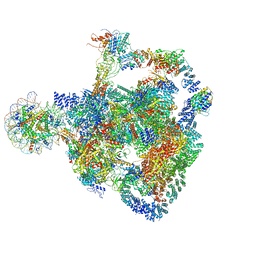 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8RC4
 
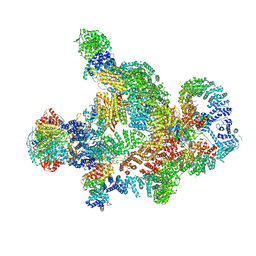 | | Structure of Integrator-PP2A complex | | Descriptor: | DSS1, Integrator complex subunit 1, Integrator complex subunit 10, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
4RW1
 
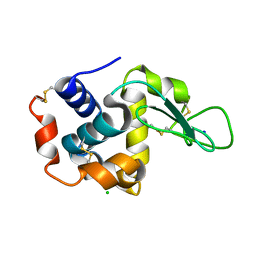 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: original beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Botha, S, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
3DBS
 
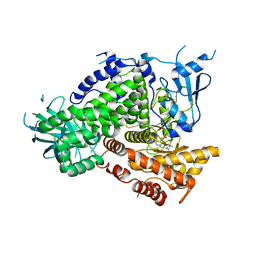 | | Structure of PI3K gamma in complex with GDC0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Wiesmann, C, Ultsch, M. | | Deposit date: | 2008-06-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer
J.Med.Chem., 51, 2008
|
|
4TOA
 
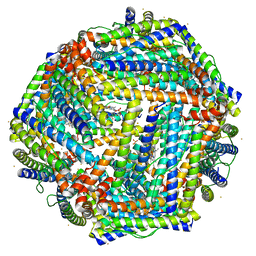 | | 1.95A resolution structure of Iron Bound BfrB (N148L) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TPG
 
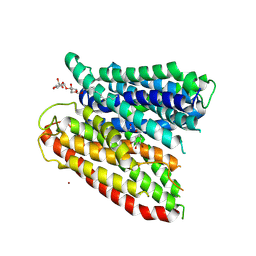 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | Descriptor: | Ala-L-3-Br-Tyr-Ala, DODECYL-BETA-D-MALTOSIDE, Proton:oligopeptide symporter POT family, ... | | Authors: | Guettou, F, Quistgaard, E.M, Raba, M, Moberg, P, Low, C, Nordlund, P. | | Deposit date: | 2014-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7UWR
 
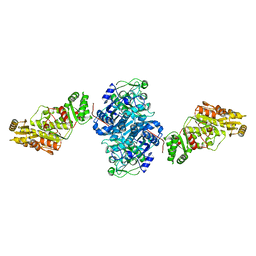 | | KSQ+AT from first module of the pikromycin synthase | | Descriptor: | Narbonolide/10-deoxymethynolide synthase PikA1, modules 1 and 2 | | Authors: | Keatinge-Clay, A.T, Dickinson, M.S, Miyazawa, T, McCool, R.S. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Priming enzymes from the pikromycin synthase reveal how assembly-line ketosynthases catalyze carbon-carbon chemistry.
Structure, 30, 2022
|
|
4TTV
 
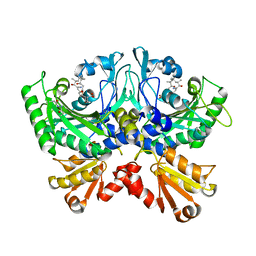 | | Crystal structure of human ThrRS complexing with a bioengineered macrolide BC194 | | Descriptor: | (1R,2R)-2-[(2S,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadec-6-en-2-yl]cyclobutanecarboxylic acid, Threonine--tRNA ligase, cytoplasmic, ... | | Authors: | Fang, P, Guo, M. | | Deposit date: | 2014-06-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor.
Sci Rep, 5, 2015
|
|
8R19
 
 | |
