8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
8INT
 
 | |
8INW
 
 | |
8INY
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir
To Be Published
|
|
8IP1
 
 | | Escherichia coli OpgD mutant-D388N with beta-1,2-glucan | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucans biosynthesis protein D, TRIETHYLENE GLYCOL, ... | | Authors: | Motouchi, S, Nakajima, M. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identification of enzymatic functions of osmo-regulated periplasmic glucan biosynthesis proteins from Escherichia coli reveals a novel glycoside hydrolase family.
Commun Biol, 6, 2023
|
|
8IU6
 
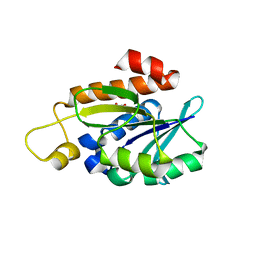 | | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Pandey, R, Tripathi, S, Lanka, A.K, Zohib, M, Pal, R.K, Arora, A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium
To Be Published
|
|
3QQR
 
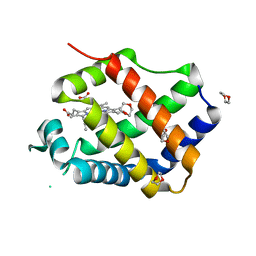 | | Crystal structure of Parasponia hemoglobin; Differential Heme Coordination is Linked to Quaternary Structure | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Non-legume hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kakar, S, Sturms, R, Savage, A, Nix, J.C, Dispirito, A, Hargrove, M.S. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structures of Parasponia and Trema hemoglobins: differential heme coordination is linked to quaternary structure.
Biochemistry, 50, 2011
|
|
3B9X
 
 | |
8J1R
 
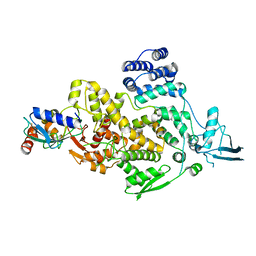 | | cryo-EM structures of Ufd4 in complex with Ubc4-Ub | | Descriptor: | Ubiquitin fusion degradation protein 4, Ubiquitin-conjugating enzyme E2 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Cai, H.Y, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Visualization of HECT-E3 Ufd4 accepting and transferring Ubiquitin to Form K29/K48-branched Polyubiquitination on N-degron. bioRxiv,doi: ttps://doi.org/10.1101/2023.05.23.542033
To Be Published
|
|
8JHH
 
 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
8J1P
 
 | | Cryo-EM structure of Ufd4 in complex with K29/48 triUb | | Descriptor: | Ubiquitin, Ubiquitin fusion degradation protein 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural Insights into the Molecular Mechanism of Ufd4-catalyzed Elongation of K48-linked Ubiquitin Chain through Lys29 Linkage
To Be Published
|
|
3QSM
 
 | | Crystal structure for the MSOX.chloride binary complex | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
8YR4
 
 | | Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2 | | Descriptor: | ATP-binding cassette sub-family B member 6, CADMIUM ION, Phytochelatin 2 | | Authors: | Choi, S.H, Lee, S.S, Lee, H.Y, Kim, S, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-03-20 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of cadmium-bound human ABCB6.
Commun Biol, 7, 2024
|
|
8YR3
 
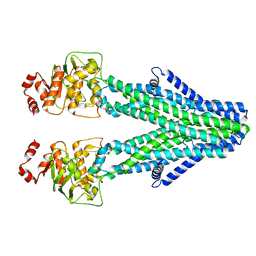 | | Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH | | Descriptor: | ATP-binding cassette sub-family B member 6, CADMIUM ION, GLUTATHIONE | | Authors: | Choi, S.H, Lee, S.S, Lee, H.Y, Kim, S, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-03-20 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of cadmium-bound human ABCB6.
Commun Biol, 7, 2024
|
|
8U5G
 
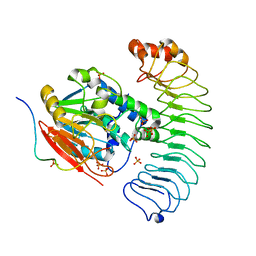 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
8V2T
 
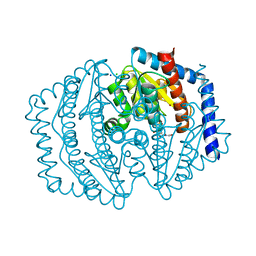 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148591 | | Descriptor: | 1,5,6-trideoxy-6,6-difluoro-1-(N-hydroxyformamido)-6-phosphono-D-ribo-hexitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
3EFZ
 
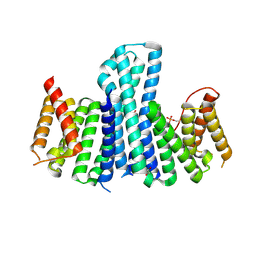 | | Crystal Structure of a 14-3-3 protein from cryptosporidium parvum (cgd1_2980) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein | | Authors: | Wernimont, A.K, Dong, A, Qiu, W, Lew, J, Wasney, G.A, Vedadi, M, Kozieradzki, I, Zhao, Y, Ren, H, Alam, Z, Lin, Y.H, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
8Z3S
 
 | | Activation mechanism and novel binding site of the BKCa channel activator CTIBD | | Descriptor: | 4-[4-(4-chlorophenyl)-3-(trifluoromethyl)-1,2-oxazol-5-yl]benzene-1,3-diol, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Lee, N, Kim, S, Jo, H, Lee, N.Y, Jin, M.S, Park, C.S. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation mechanism and novel binding sites of the BK Ca channel activator CTIBD.
Life Sci Alliance, 7, 2024
|
|
5ZHZ
 
 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | Descriptor: | Probable endonuclease 4, SULFATE ION, ZINC ION | | Authors: | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
8V4J
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
6APB
 
 | |
6AQJ
 
 | | Crystal structures of Staphylococcus aureus ketol-acid reductoisomerase in complex with two transition state analogs that have biocidal activity. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Gracia, M, Lv, Y, Schembri, M.A, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.373 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
7EY3
 
 | | Double cysteine mutations in T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hamdan, S.H, Leow, T.C, Yahaya, N.M, Ali, M.S.M, Jonet, M.A, Mohamad Aris, S.N.A, Maiangwa, J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Knotting terminal ends of mutant T1 lipase with disulfide bond improved structure rigidity and stability.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
5Z9H
 
 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
7F3V
 
 | | Crystal structure of YfiH with C107A mutation in complex with endogenous UDP-MurNAc | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Lee, M.S, Hsieh, K.Y, Chang, C.I. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for the Peptidoglycan-Editing Activity of YfiH.
Mbio, 13, 2021
|
|
