6EO0
 
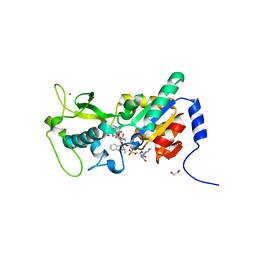 | | Zebrafish Sirt5 in complex with stalled peptidylimidate and bicyclic intermediate of inhibitory compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[(2~{S},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2-[[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]amino]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]amino]propanoic acid, 3-[[(~{Z})-~{C}-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]sulfanyl-~{N}-[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]carbonimidoyl]amino]propanoic acid, ... | | Authors: | Pannek, M, Steegborn, C. | | Deposit date: | 2017-10-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5X1A
 
 | |
5X1D
 
 | |
2QC3
 
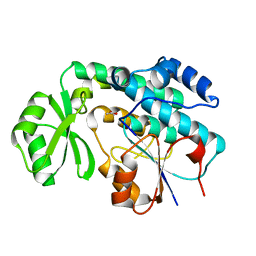 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
6ER7
 
 | | CHEMOTAXIS PROTEIN CHEY FROM Pyrococcus horikoshiI | | Descriptor: | 120aa long hypothetical chemotaxis protein (CheY) | | Authors: | Paithankar, K.S, Enderle, M.E, Wirthensohn, D, Grininger, M, Oesterhelt, D. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the archaeal chemotaxis protein CheY in a domain-swapped dimeric conformation.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1QO4
 
 | | ARABIDOPSIS THALIANA PEROXIDASE A2 AT ROOM TEMPERATURE | | Descriptor: | CALCIUM ION, PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Henriksen, A, Gajhede, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Activity and Structure of Highly Similar Peroxidases. Spectroscopic, Crystallographic, and Enzymatic Analyses of Lignifying Arabidopsis Thaliana Peroxidase A2 and Horseradish Peroxidase A2
Biochemistry, 40, 2001
|
|
6EL4
 
 | |
1R77
 
 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | Descriptor: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | Authors: | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | Deposit date: | 2003-10-20 | | Release date: | 2005-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
2QO5
 
 | | Crystal structure of the cysteine 91 threonine mutant of zebrafish liver bile acid-binding protein complexed with cholic acid | | Descriptor: | CHOLIC ACID, Liver-basic fatty acid binding protein | | Authors: | Capaldi, S, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2007-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Single Amino Acid Mutation in Zebrafish (Danio rerio) Liver Bile Acid-binding Protein Can Change the Stoichiometry of Ligand Binding.
J.Biol.Chem., 282, 2007
|
|
4ALC
 
 | | X-Ray photoreduction of Polysaccharide monooxigenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
6EKY
 
 | |
6M7L
 
 | | Complex of OxyA with the X-domain from GPA biosynthesis | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase, Putative non-ribosomal peptide synthetase | | Authors: | Greule, A, Izore, T, Tailhades, J, Peschke, M, Schoppet, M, Ahmed, I, Kulik, A, Adamek, M, Ziemert, N, De Voss, J, Stegmann, E, Cryle, M.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.648297 Å) | | Cite: | Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Nat Commun, 10, 2019
|
|
5WWP
 
 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
6EHB
 
 | |
6ENX
 
 | | Zebrafish Sirt5 in complex with stalled bicyclic intermediate of inhibitory compound 10 | | Descriptor: | 4-[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2-[[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]amino]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]butanoic acid, DIMETHYL SULFOXIDE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Pannek, M, Steegborn, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8JV7
 
 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPADS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-[4-methanoyl-6-methyl-5-oxidanyl-3-(phosphonooxymethyl)pyridin-2-yl]diazenyl]benzene-1,3-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
8JV8
 
 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPNDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
1Z4U
 
 | | hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00799585 | | Descriptor: | (2Z)-2-[(1-ADAMANTYLCARBONYL)AMINO]-3-[4-(2-BROMOPHENOXY)PHENYL]PROP-2-ENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M, Nugent, R, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R. | | Deposit date: | 2005-03-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
3ZKM
 
 | | BACE2 FAB COMPLEX | | Descriptor: | BETA-SECRETASE 2, DIMETHYL SULFOXIDE, FAB HEAVY CHAIN, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6L5P
 
 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | GgCGT, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
3GA4
 
 | | Crystal structure of Ost6L (photoreduced form) | | Descriptor: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6ZJR
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
4LZS
 
 | | Crystal Structure of BRD4(1) bound to inhibitor XD46 | | Descriptor: | 4-acetyl-3-ethyl-N,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Einsle, O, Gerhardt, S. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6TP3
 
 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
