5E1Z
 
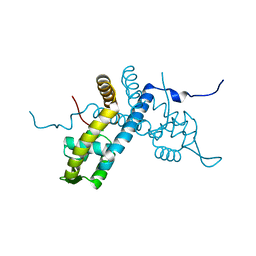 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,4-dichlorophenol bound form | | Descriptor: | 2,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
to be published
|
|
5E20
 
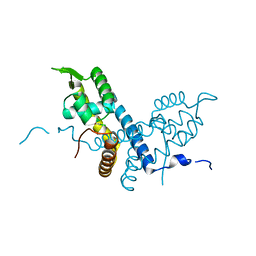 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,3-dichlorophenol bound form | | Descriptor: | 2,3-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
To be published
|
|
5NW2
 
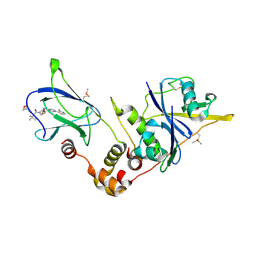 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-3,3-dimethyl-2-(oxetane-3-carboxamido)butanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 19) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(oxetan-3-ylcarbonylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
2Q1L
 
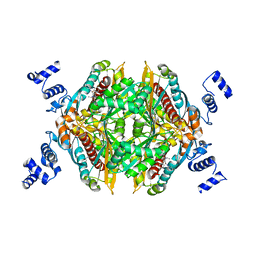 | | Design and Synthesis of Pyrrole-based, Hepatoselective HMG-CoA Reductase Inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-05-24 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and synthesis of hepatoselective, pyrrole-based HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5LNE
 
 | | E. coli F9 pilus adhesin FmlH bound to the Thomsen-Friedenreich (TF) antigen | | Descriptor: | NICKEL (II) ION, Putative Fml fimbrial adhesin FmlD, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Ruer, S, Conover, M.S, Kalas, V, Taganna, J, De Greve, H, Pinkner, J.S, Dodson, K.W, Hultgren, S.J, Remaut, H. | | Deposit date: | 2016-08-04 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection.
Cell Host Microbe, 20, 2016
|
|
6A1C
 
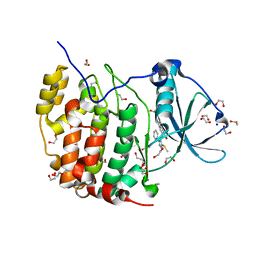 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
1X76
 
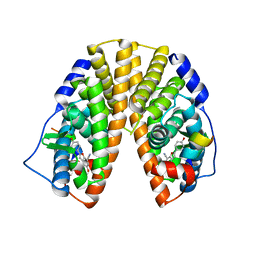 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-697 | | Descriptor: | 5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-CARBONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X7E
 
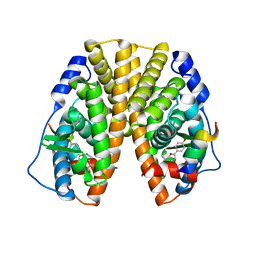 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor 1 (alpha), [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1XPX
 
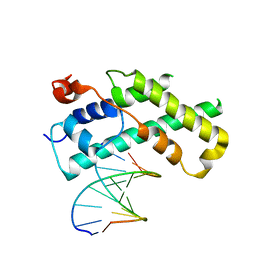 | |
5D6S
 
 | | Structure of epoxyqueuosine reductase from Streptococcus thermophilus. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER | | Authors: | Payne, K.A.P, Fisher, K, Dunstan, M.S, Sjuts, H, Leys, D. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
J.Biol.Chem., 290, 2015
|
|
2PJR
 
 | | HELICASE PRODUCT COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
6R7W
 
 | | Tannerella forsythia mature mirolysin in complex with a cleaved peptide. | | Descriptor: | CALCIUM ION, CITRIC ACID, ETHANOL, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
5DC5
 
 | | Crystal structure of D176N HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
1XOY
 
 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | Descriptor: | hypothetical protein At3g04780.1 | | Authors: | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
6OK1
 
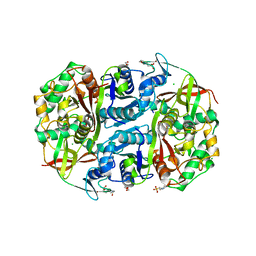 | | Ltp2-ChsH2(DUF35) aldolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ChsH2(DUF35), ... | | Authors: | Kimber, M.S, Mallette, E, Aggett, R, Seah, S.Y.K. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35is a thiolase superfamily member with a radically repurposed active site.
J.Biol.Chem., 294, 2019
|
|
6OS8
 
 | |
6OU2
 
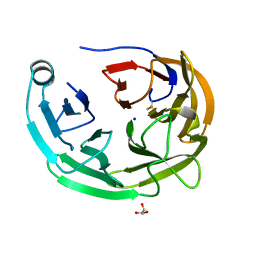 | |
1UVO
 
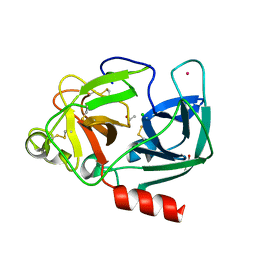 | | Structure Of The Complex Of Porcine Pancreatic Elastase In Complex With Cadmium Refined At 1.85 A Resolution (Crystal A) | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Mueller-Dieckmann, C, Tucker, P.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the Influence of the Incident Photon Energy on the Radiation Damage in Crystalline Biological Samples
J.Synchrotron Radiat., 12, 2005
|
|
6AEX
 
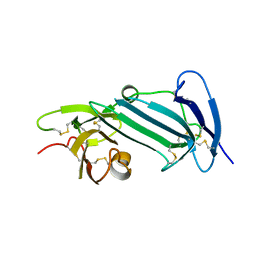 | | Crystal structure of unoccupied murine uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-08-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of the unoccupied murine urokinase-type plasminogen activator receptor (uPAR) reveals a tightly packed DII-DIII unit.
Febs Lett., 593, 2019
|
|
6OBU
 
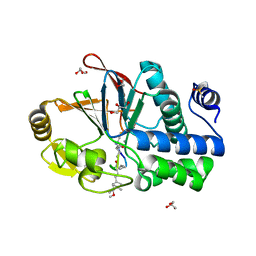 | | PP1 Y134K in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2V2V
 
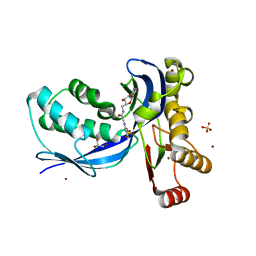 | | IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, 5'-[(1H-BENZIMIDAZOL-2-YLACETYL)AMINO]-5'-DEOXYCYTIDINE, BROMIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Characterization of Cytidine Derivatives that Inhibit the Kinase Ispe of the Non-Mevalonate Pathway for Isoprenoid Biosynthesis.
Chemmedchem, 3, 2008
|
|
5ZA7
 
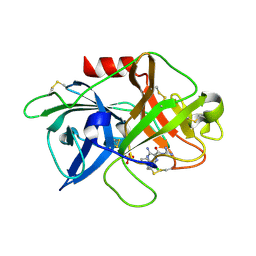 | | uPA-HMA | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-[bis(azanyl)methylidene]-6-chloranyl-pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
5ZAE
 
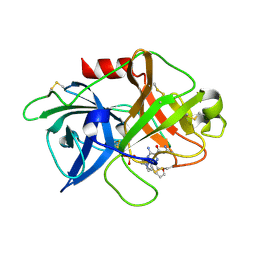 | | uPA-6F-HMA | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
5ZC5
 
 | | uPA-NU-09F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(4-fluoranyl-1-benzofuran-2-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Buckley, B.J, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
8TKQ
 
 | |
