2V75
 
 | | N-terminal domain of Nab2 | | Descriptor: | NUCLEAR POLYADENYLATED RNA-BINDING PROTEIN NAB2 | | Authors: | Grant, R.P, Marshall, N.J, Stewart, M. | | Deposit date: | 2007-07-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-Terminal Mlp1-Binding Domain of the Saccharomyces Cerevisiae Mrna-Binding Protein, Nab2.
J.Mol.Biol., 376, 2008
|
|
2VDV
 
 | | Structure of trm8, m7G methylation enzyme | | Descriptor: | S-ADENOSYLMETHIONINE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
8AGI
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
3IQJ
 
 | | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (10mer) | | Descriptor: | 10-mer peptide from RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Ottmann, C, Weyand, M. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol.Cell.Biol., 30, 2010
|
|
1Z6F
 
 | | Crystal structure of penicillin-binding protein 5 from E. coli in complex with a boronic acid inhibitor | | Descriptor: | GLYCEROL, N1-[(1R)-1-(DIHYDROXYBORYL)ETHYL]-N2-[(TERT-BUTOXYCARBONYL)-D-GAMMA-GLUTAMYL]-N6-[(BENZYLOXY)CARBONYL-L-LYSINAMIDE, Penicillin-binding protein 5 | | Authors: | Nicola, G, Peddi, S, Stefanova, M, Nicholas, R.A, Gutheil, W.G, Davies, C. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli Penicillin-Binding Protein 5 Bound to a Tripeptide Boronic Acid Inhibitor: A Role for Ser-110 in Deacylation.
Biochemistry, 44, 2005
|
|
1ZAX
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form B | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
1ZBY
 
 | | High-Resolution Crystal Structure of Native (Resting) Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | Deposit date: | 2005-04-09 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
3NWN
 
 | | Crystal structure of the human KIF9 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF9, ... | | Authors: | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human KIF9 motor domain in complex with ADP
To be Published
|
|
5A8M
 
 | | Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
3VOR
 
 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2UV4
 
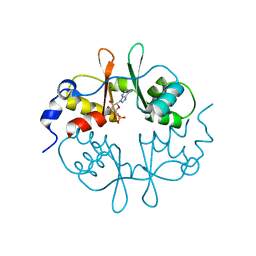 | | Crystal Structure of a CBS domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ADENOSINE MONOPHOSPHATE | | Authors: | Day, P, Sharff, A, PArra, L, Cleasby, A, Williams, M, Horer, S, Nar, H, Redemann, N, Tickle, I, Yon, J. | | Deposit date: | 2007-03-09 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure of a Cbs-Domain Pair from the Regulatory Gamma1 Subunit of Human Ampk in Complex with AMP and Zmp.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4HT7
 
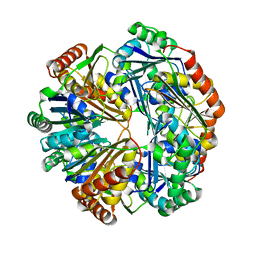 | | CO2 concentrating mechanism protein P, CcmP form 2 | | Descriptor: | CO2 concentrating mechanism protein P | | Authors: | Cai, F, Sutter, M, Kerfeld, C.A. | | Deposit date: | 2012-10-31 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment.
J.Biol.Chem., 288, 2013
|
|
1ZCD
 
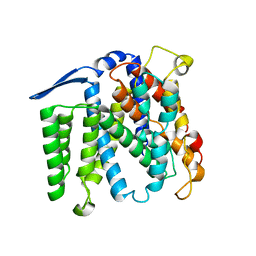 | | Crystal structure of the Na+/H+ antiporter NhaA | | Descriptor: | Na(+)/H(+) antiporter 1 | | Authors: | Hunte, C, Screpanti, E, Venturi, M, Rimon, A, Padan, E, Michel, H. | | Deposit date: | 2005-04-11 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH.
Nature, 435, 2005
|
|
3OAH
 
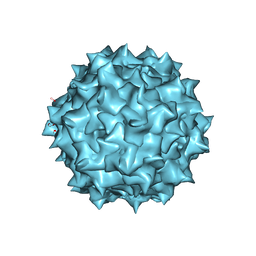 | |
2V3S
 
 | | Structural insights into the recognition of substrates and activators by the OSR1 kinase | | Descriptor: | ACETATE ION, SERINE/THREONINE-PROTEIN KINASE OSR1, SERINE/THREONINE-PROTEIN KINASE WNK4 | | Authors: | Villa, F, Goebel, J, Rafiqi, F.H, Deak, M, Thastrup, J, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2007-06-21 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Recognition of Substrates and Activators by the Osr1 Kinase
Embo Rep., 8, 2007
|
|
4HXT
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR329 | | Descriptor: | De Novo Protein OR329 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, X, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-12 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
3VQS
 
 | | Crystal structure of HCV NS5B RNA polymerase with a novel piperazine inhibitor | | Descriptor: | (2R)-4-(5-cyclopropyl[1,3]thiazolo[4,5-d]pyrimidin-2-yl)-N-[3-fluoro-4-(trifluoromethoxy)benzyl]-1-{[4-(trifluoromethyl)phenyl]sulfonyl}piperazine-2-carboxamide, CHLORIDE ION, RNA-directed RNA polymerase | | Authors: | Adachi, T, Doi, S, Ando, I, Sugimoto, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preclinical Characterization of JTK-853, a Novel Nonnucleoside Inhibitor of the Hepatitis C Virus RNA-Dependent RNA Polymerase.
Antimicrob.Agents Chemother., 56, 2012
|
|
8AGJ
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112A in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
1ZDQ
 
 | | Crystal Structure of Met150Gly AfNiR with Methylsulfanyl Methane Bound | | Descriptor: | (METHYLSULFANYL)METHANE, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, MacPherson, I.S, Alexandre, M, Diederix, R.E.M, Canters, G.W, Murphy, M.E.P, Verbeet, M.P. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A rearranging ligand enables allosteric control of catalytic activity in copper-containing nitrite reductase.
J.Mol.Biol., 358, 2006
|
|
1ZEY
 
 | | CGG A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*GP*CP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8AGL
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
1Z6U
 
 | | Np95-like ring finger protein isoform b [Homo sapiens] | | Descriptor: | Np95-like ring finger protein isoform b, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Crystal Structure of the Human Ubiquitin Liagse NIRF
To be Published
|
|
2UZ3
 
 | | Crystal Structure of Thymidine Kinase with dTTP from U. urealyticum | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2007-04-24 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Thymidine Kinase 1 of Human and Mycoplasmic Origin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Z6Y
 
 | | Structure Of Human ADP-Ribosylation Factor-Like 5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Of Human ADP-Ribosylation Factor-Like 5
To be Published
|
|
