1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YOK
 
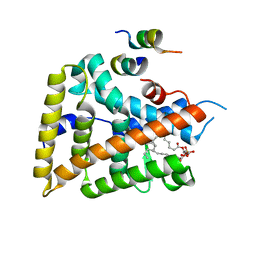 | | crystal structure of human LRH-1 bound with TIF-2 peptide and phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, MacKay, J.A, Juzumiene, D, Bynum, J.M, Madauss, K, Montana, V, Lebedeva, L, Suzawa, M, Williams, J.D, Williams, S.P, Guy, R.K, Thornton, J.W, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1.
Cell(Cambridge,Mass.), 120, 2005
|
|
1YSX
 
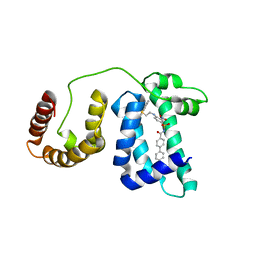 | | Solution structure of domain 3 from human serum albumin complexed to an anti-apoptotic ligand directed against Bcl-xL and Bcl-2 | | Descriptor: | 4-({2-[(2,4-DIMETHYLPHENYL)SULFANYL]ETHYL}AMINO)-N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITROBENZENESULFONAMIDE, Serum albumin | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
7ZQI
 
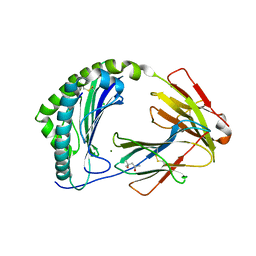 | | MHC class I from a wild bird in complex with a nonameric peptide P2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
3NTX
 
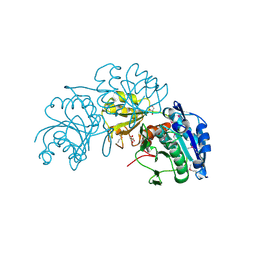 | | Crystal Structure of L-asparaginase I from Yersinia pestis | | Descriptor: | Cytoplasmic L-asparaginase I, GLYCEROL | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-05 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L-asparaginase I from Yersinia pestis
To be Published, 2010
|
|
4ZVI
 
 | | GYRASE B IN COMPLEX WITH 4,5-DIBROMOPYRROLAMIDE-BASED INHIBITOR | | Descriptor: | DNA gyrase subunit B, IODIDE ION, N-(4-{[(4,5-dibromo-1H-pyrrol-2-yl)carbonyl]amino}benzoyl)glycine | | Authors: | Zidar, N, Macut, H, Tomasic, T, Brvar, M, Montalvao, S, Tammela, P, Solmajer, T, Peterlin Masic, L, Ilas, J, Kikelj, D. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Phenyl-4,5-dibromopyrrolamides and N-Phenylindolamides as ATP Competitive DNA Gyrase B Inhibitors: Design, Synthesis, and Evaluation.
J.Med.Chem., 58, 2015
|
|
2RRM
 
 | | Interplay between phosphatidyl-inositol-phosphates and claudins upon binding to the 1st PDZ domain of zonula occludens 1 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Hiroaki, H, Satomura, K, Goda, N, Umetsu, Y, Taniguchi, R, Ikegami, T, Furuse, M. | | Deposit date: | 2011-01-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, and 15N resonance assignment of the first PDZ domain of mouse ZO-1
Biomol.Nmr Assign., 5, 2011
|
|
1YMM
 
 | | TCR/HLA-DR2b/MBP-peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Hahn, M, Nicholson, M.J, Pyrdol, J, Wucherpfennig, K.W. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor.
NAT.IMMUNOL., 6, 2005
|
|
7ZQJ
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P3 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
4HLM
 
 | | Crystal structure of Tankyrase 2 in complex with 3',4'-Dihydroxyflavone | | Descriptor: | 2-(3,4-dihydroxyphenyl)-4H-chromen-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
3NXZ
 
 | | Crystal Structure of UreE from Helicobacter pylori (Cu2+ bound form) | | Descriptor: | COPPER (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
2RQ9
 
 | | Solution structure of human acidic fibroblast growth factor (aFGF) in the presence of a protein stabilizer NDSB-new | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Enomoto, M, Xiang, L, Ishii, T, Tochio, N, Hosoda, K, Inoue, Y, Nameki, N, Wakamatsu, K. | | Deposit date: | 2009-03-12 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human acidic fibroblast growth factor (aFGF) in the presence of a protein stabilizer NDSB-new
To be Published
|
|
2RT3
 
 | | Solution structure of the second RRM domain of Nrd1 | | Descriptor: | Negative regulator of differentiation 1 | | Authors: | Kobayashi, A, Kanaba, T, Mishima, M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the second RRM domain of Nrd1, a fission yeast MAPK target RNA binding protein, and implication for its RNA recognition and regulation
Biochem.Biophys.Res.Commun., 437, 2013
|
|
3O39
 
 | |
4HTL
 
 | | Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Beta-glucoside kinase | | Authors: | Osipiuk, J, Mack, J, Endres, M, Salazar, J, Zhang, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes.
To be Published
|
|
1YT9
 
 | | HIV Protease with oximinoarylsulfonamide bound | | Descriptor: | (S)-N-((2S,3R)-3-HYDROXY-4-(4-((E)-(HYDROXYIMINO)METHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YL)-3-METHYL-2-(3 -((2-METHYLTHIAZOL-4-YL)METHYL)-2-OXOIMIDAZOLIDIN-1-YL)BUTANAMIDE, Pol polyprotein | | Authors: | Yeung, C.M, Klein, L.L, Flentge, C.A, Randolph, J.T, Zhao, C, Sun, M, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | Deposit date: | 2005-02-10 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oximinoarylsulfonamides as potent HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3O3H
 
 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and manganese ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
4HHR
 
 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
2RUK
 
 | |
1YUO
 
 | | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG)from atlantic cod (Gadus morhua) | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Moe, E, Leiros, I, Riise, E.K, Olufsen, M, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua)
J.Mol.Biol., 343, 2004
|
|
1Z40
 
 | | AMA1 from Plasmodium falciparum | | Descriptor: | CHLORIDE ION, apical membrane antigen 1 precursor | | Authors: | Bai, T, Becker, M, Gupta, A, Strike, P, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2005-03-14 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3O4A
 
 | |
4HU4
 
 | | Crystal structure of EAL domain of the E. coli DosP - dimeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ZMD
 
 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD+/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
4ZVF
 
 | | Crystal structure of GGDEF domain of the E. coli DosC - form II (GTP-alpha-S-bound) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Diguanylate cyclase DosC, ... | | Authors: | Tarnawski, M, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural analysis of an oxygen-regulated diguanylate cyclase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
