1A53
 
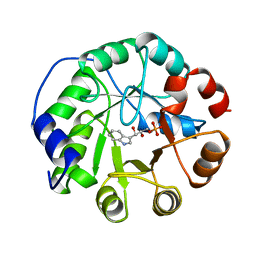 | | COMPLEX OF INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS WITH INDOLE-3-GLYCEROLPHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1998-02-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
J.Mol.Biol., 319, 2002
|
|
7ZIR
 
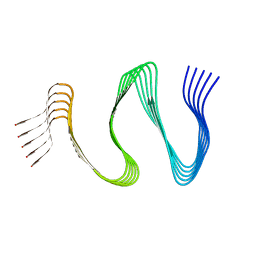 | | Cryo-EM structure of hnRNPDL amyloid fibrils | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D-like | | Authors: | Garcia-Pardo, J, Chaves-Sanjuan, A, Bartolome-Nafria, A, Gil-Garcia, M, Visentin, C, Bolognesi, M, Ricagno, S, Ventura, S. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3.
Nat Commun, 14, 2023
|
|
1RD5
 
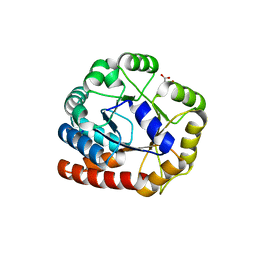 | | Crystal structure of Tryptophan synthase alpha chain homolog BX1: a member of the chemical plant defense system | | Descriptor: | MALONIC ACID, Tryptophan synthase alpha chain, chloroplast | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
7QNN
 
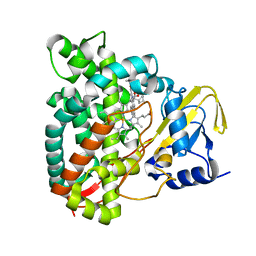 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclopentylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
4MOU
 
 | | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282 | | Descriptor: | CACODYLATE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Chapman, H.C, Cooper, D.R, Geffken, K.T, Cymborowski, M.T, Osinski, T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282
To be Published
|
|
8FUP
 
 | | Bromodomain of CBP liganded with BMS-536924 and CCS-1477 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
7ZZ6
 
 | | Cryo-EM structure of "CT-CT dimer" of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
8G0N
 
 | |
3MF2
 
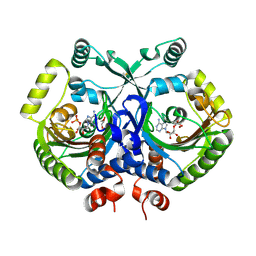 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bll0957 protein, GLYCEROL, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8FXN
 
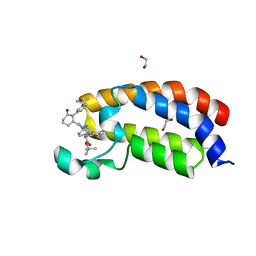 | | Bromodomain of CBP liganded with iCBP7 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, tert-butyl {(1R,4s)-4-[(5M)-2-[(2S)-1-(3-tert-butylphenyl)-6-oxopiperidin-2-yl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-benzimidazol-1-yl]cyclohexyl}carbamate | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks are sensitive to EP300/CBP bromodomain inhibition
To be published
|
|
4MSU
 
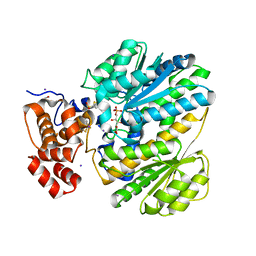 | | Human GKRP bound to AMG-6861 and Sorbitol-6-phosphate | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-{4-[4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Ashton, K.S, Andrews, K.L, Bryan, M.C, Chen, J, Chen, K, Chen, M, Chmait, S, Croghan, M, Cupples, R, Fotsch, C, Helmering, J, Jordan, S.R, Kurzeja, R.J, Michelsen, K, Pennington, L.D, Poon, S.F, Sivits, G, Van, G, Vonderfecht, S.L, Wahl, R.C, Zhang, J, Lloyd, D.J, Hale, C, St Jean, D.J. | | Deposit date: | 2013-09-18 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 1. Discovery of a Novel Tool Compound for in Vivo Proof-of-Concept.
J.Med.Chem., 57, 2014
|
|
7LKK
 
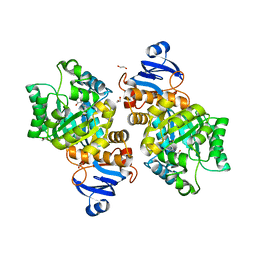 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) in complex with Methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 1,2-ETHANEDIOL, Aminofutalosine deaminase, ... | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
4MVN
 
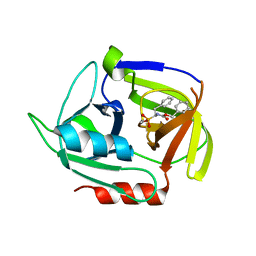 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
8G1H
 
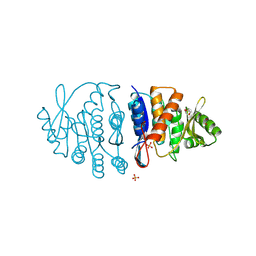 | | Ancestral protein AncTh of Phosphomethylpirimidine kinases family | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphomethylpyrimidine Kinase | | Authors: | Perez, M, Munoz, S, Cea, P, Castro-Fernandez, V. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deciphering Structural Traits for Thermal and Kinetic Stability across Protein Family Evolution through Ancestral Sequence Reconstruction.
Mol.Biol.Evol., 41, 2024
|
|
6TGJ
 
 | |
6HBN
 
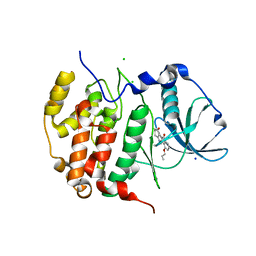 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA/CSKN2A1 GENE PRODUCT) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Hochscherf, J, Dimper, V, Witulski, B, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
3MH3
 
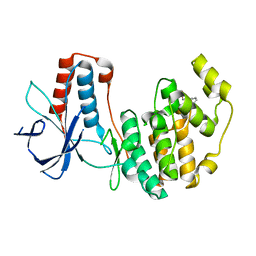 | | Mutagenesis of p38 MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-out state | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M, Bukhtiyarova, M, Springman, E.B. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis of p38alpha MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-OUT state.
Biochemistry, 46, 2007
|
|
6TGV
 
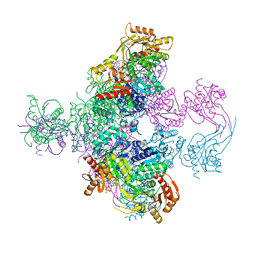 | | Crystal structure of Mycobacterium smegmatis CoaBC in complex with CTP and FMN | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein CoaBC, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
5A36
 
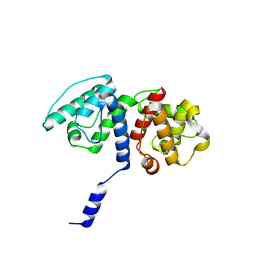 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
6HG7
 
 | | Crystal structure of a collagen II fragment containing the binding site of PEDF and COMP, (POG)4-LKG HRG FTG LQG-POG(4) | | Descriptor: | Collagen alpha-1(II) chain, SULFATE ION | | Authors: | Gebauer, J.M, Koehler, A, Dietmar, H, Gompert, M, Neundorf, I, Zaucke, F, Koch, M, Baumann, U. | | Deposit date: | 2018-08-22 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | COMP and TSP-4 interact specifically with the novel GXKGHR motif only found in fibrillar collagens.
Sci Rep, 8, 2018
|
|
7F44
 
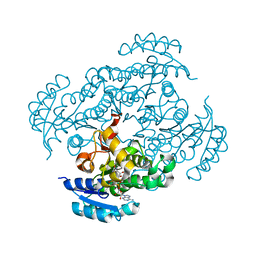 | | Crystal structure of Moraxella catarrhalis enoyl-ACP-reductase (FabI) in complex with the cofactor NAD | | Descriptor: | CALCIUM ION, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Katiki, M, Neetu, N, Pratap, S, Kumar, P. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Biochemical and structural basis for Moraxella catarrhalis enoyl-acyl carrier protein reductase (FabI) inhibition by triclosan and estradiol.
Biochimie, 198, 2022
|
|
2N7D
 
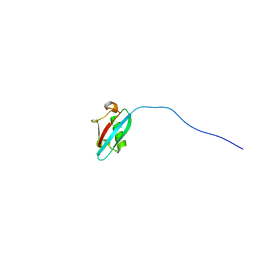 | |
8FTQ
 
 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
4ZZA
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose, selenomethionine derivative | | Descriptor: | Sugar binding protein of ABC transporter system, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2015-05-22 | | Release date: | 2016-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
6HCK
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with dipeptide Leu-Leu | | Descriptor: | LEUCINE, Listeriolysin regulatory protein, SODIUM ION | | Authors: | Grundstrom, C, Oelker, M, Krypotou, E, Scortti, M, Luisi, B.F, Vazquez-Boland, J, Sauer-Eriksson, A.E. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of Bacterial Virulence through the Peptide Signature of the Habitat.
Cell Rep, 26, 2019
|
|
