7Y0I
 
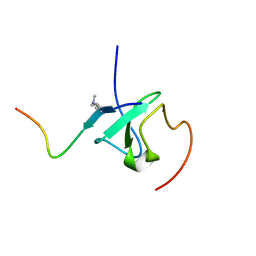 | | Solution structures of ASH1L PHD domain in complex with H3K4me2 peptide | | Descriptor: | ALA-ARG-THR-MLY-GLN-THR-ALA-ARG-LYS-SER-THR-GLY-GLY-LYS-ALA, Histone-lysine N-methyltransferase ASH1L, ZINC ION | | Authors: | Yu, M, Zeng, L. | | Deposit date: | 2022-06-05 | | Release date: | 2022-10-12 | | Method: | SOLUTION NMR | | Cite: | Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer.
Front Oncol, 12, 2022
|
|
7KKF
 
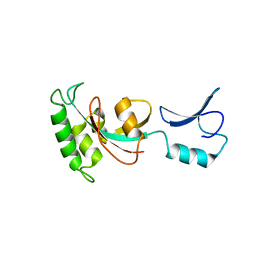 | |
7XZX
 
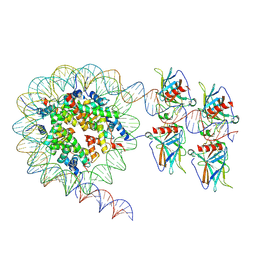 | | Cryo-EM structure of the nucleosome in complex with p53 DNA-binding domain | | Descriptor: | Cellular tumor antigen p53, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
3B9B
 
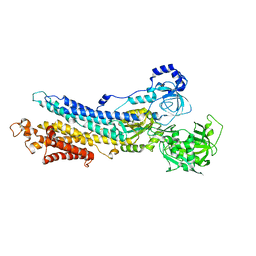 | | Structure of the E2 beryllium fluoride complex of the SERCA Ca2+-ATPase | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Olesen, C, Picard, M, Winther, A.M.L, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structural basis of calcium transport by the calcium pump.
Nature, 450, 2007
|
|
7XT3
 
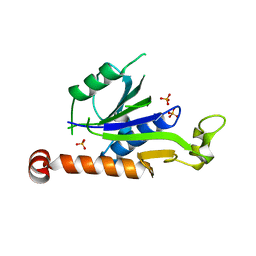 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
7F26
 
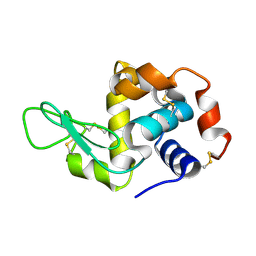 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Liang, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3BDN
 
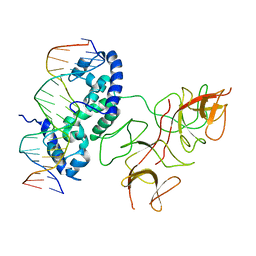 | | Crystal Structure of the Lambda Repressor | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DAP*DCP*DCP*DAP*DCP*DTP*DGP*DGP*DCP*DGP*DGP*DTP*DGP*DAP*DTP*DAP*DT)-3'), DNA (5'-D(*DTP*DAP*DTP*DAP*DTP*DCP*DAP*DCP*DCP*DGP*DCP*DCP*DAP*DGP*DTP*DGP*DGP*DTP*DAP*DT)-3'), Lambda Repressor | | Authors: | Stayrook, S.E, Jaru-Ampornpan, P, Hochschild, A, Lewis, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.909 Å) | | Cite: | Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding
Nature, 452, 2008
|
|
3BF0
 
 | |
7EQ9
 
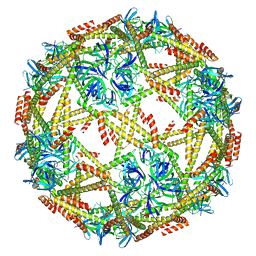 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | Descriptor: | TIP60 | | Authors: | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
3BFO
 
 | | Crystal structure of Ig-like C2-type 2 domain of the human Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (Isoform 2) | | Authors: | Walker, J.R, Akutsu, M, Littler, D.R, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of the Ig-like C2-type 2 domain of the human Mucosa-associated lymphoid tissue lymphoma translocation protein 1.
To be Published
|
|
7Y4K
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-ornitine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-5-(phenylmethoxycarbonylamino)pentanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
3B08
 
 | | Crystal structure of the mouse HOIL1-L-NZF in complex with linear di-ubiquitin | | Descriptor: | Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION, ... | | Authors: | Sato, Y, Fujita, H, Yoshikawa, A, Yamashita, M, Yamagata, A, Kaiser, S.E, Iwai, K, Fukai, S. | | Deposit date: | 2011-06-07 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BIS
 
 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6IN5
 
 | |
3EL0
 
 | |
3EOX
 
 | |
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
6IM0
 
 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | BICARBONATE ION, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
3EPM
 
 | | Crystal structure of Caulobacter crescentus ThiC | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SULFATE ION, Thiamine biosynthesis protein thiC, ... | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
3EOE
 
 | | Crystal Structure of Pyruvate Kinase from toxoplasma gondii, 55.m00007 | | Descriptor: | GLYCEROL, Pyruvate kinase | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Wasney, G, Hassani, A, Vedadi, M, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structure of Toxoplasma gondii pyruvate kinase 1.
Plos One, 5, 2010
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
3EC5
 
 | | The crystal structure of Thioflavin-T (ThT) binding OspA mutant | | Descriptor: | Outer Surface Protein A, TETRAETHYLENE GLYCOL | | Authors: | Biancalana, M, Makabe, K, Koide, A, Koide, S. | | Deposit date: | 2008-08-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies
J.Mol.Biol., 385, 2009
|
|
7Y4M
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-lysine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-6-(phenylmethoxycarbonylamino)hexanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
