6J9Y
 
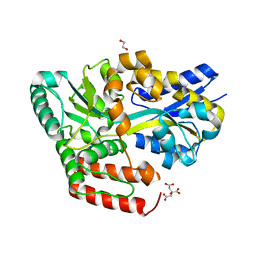 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
1JRV
 
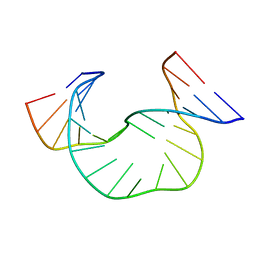 | | SOLUTION STRUCTURE OF DAATAA DNA BULGE | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
5B81
 
 | |
4LAY
 
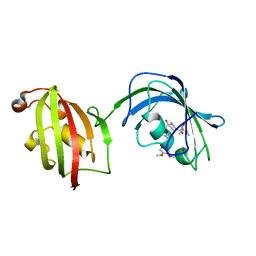 | | Crystal Structure Analysis of FKBP52, Complex with I63 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
6JP2
 
 | |
6JAG
 
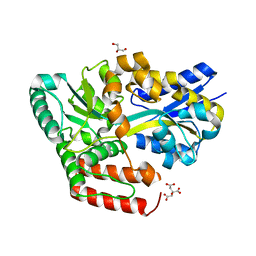 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with sucrose | | Descriptor: | ABC transporter, periplasmic substrate-binding protein, CITRIC ACID, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
7YWR
 
 | |
1JT1
 
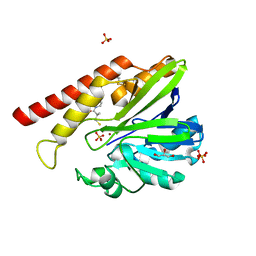 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, CHLORIDE ION, FEZ-1, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2001-08-20 | | Release date: | 2003-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-[beta]-lactamase from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.MOL.BIOL., 325, 2003
|
|
6JB4
 
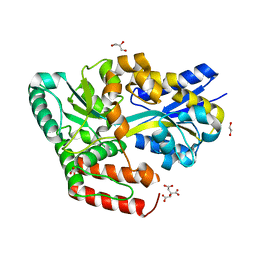 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein W287A in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6F97
 
 | | Crystal structure of the V465T mutant of 5-(Hydroxymethyl)furfural Oxidase (HMFO) | | Descriptor: | 5-(hydroxymethyl)furfural oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pickl, M, Swoboda, A, Romero, E, Winkler, C.K, Binda, C, Mattevi, A, Faber, K, Fraaije, M.W. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic Resolution of sec-Thiols by Enantioselective Oxidation with Rationally Engineered 5-(Hydroxymethyl)furfural Oxidase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2MET
 
 | | NMR spatial structure of the trimeric mutant TM domain of VEGFR2 receptor. | | Descriptor: | Vascular endothelial growth factor receptor 2 | | Authors: | Mineev, K.S, Arseniev, A.A, Shulepko, M, Lyukmanova, E.N, Kirpichnikov, M.P. | | Deposit date: | 2013-10-02 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Structure, 22, 2014
|
|
1JTC
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND LEU 44 REPLACED BY PHE (L44F) | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
2C4J
 
 | | Human glutathione-S-transferase M2-2 T210S mutant in complex with glutathione-styrene oxide conjugate | | Descriptor: | GLUTATHIONE S-TRANSFERASE MU 2, L-GAMMA-GLUTAMYL-S-[(2S)-2-HYDROXY-2-PHENYLETHYL]-L-CYSTEINYLGLYCINE | | Authors: | Tars, K, Andersson, M, Ivarsson, Y, Olin, B, Mannervik, B. | | Deposit date: | 2005-10-20 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Alternative Mutations of a Positively Selected Residue Elicit Gain or Loss of Functionalities in Enzyme Evolution.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
4LDC
 
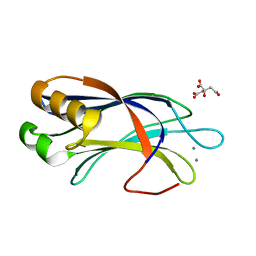 | | Crystal Structure of DOC2B C2B domain | | Descriptor: | CALCIUM ION, CITRATE ANION, Double C2-like domain-containing protein beta | | Authors: | Giladi, M, Almagor, L, Hirsch, J.A. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.264 Å) | | Cite: | The C2B Domain Is the Primary Ca(2+) Sensor in DOC2B: A Structural and Functional Analysis.
J.Mol.Biol., 425, 2013
|
|
7YVW
 
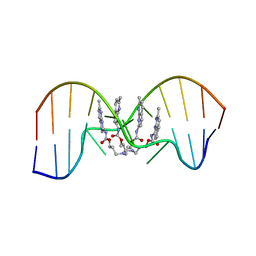 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
6ETM
 
 | |
4LH5
 
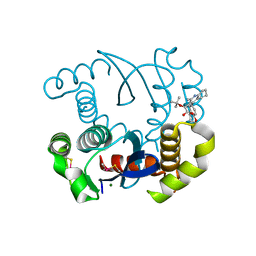 | | Dual inhibition of HIV-1 replication by Integrase-LEDGF allosteric inhibitors is predominant at post-integration stage during virus production rather than at integration | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase, MAGNESIUM ION | | Authors: | Ruff, M, Levy, N, Eiler, S. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage.
Retrovirology, 10, 2013
|
|
1JWJ
 
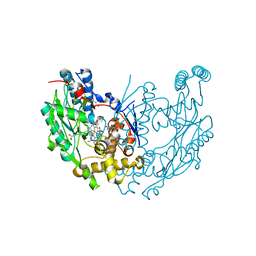 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457F Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
7YTF
 
 | | Structure of OCPx2 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein, beta,beta-carotene-4,4'-dione | | Authors: | Yang, Y.W, Chen, S.Z, Liu, K, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
7YTH
 
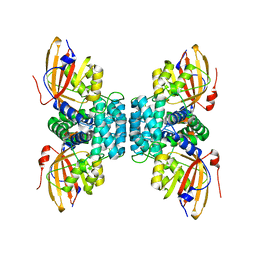 | | Structure of OCPx1 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein | | Authors: | Yang, Y.W, Liu, K, Chen, S.Z, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
6ESU
 
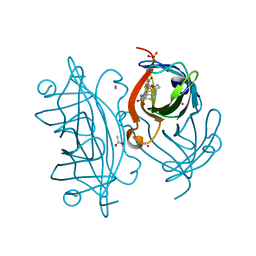 | | Artificial imine reductase mutant S112A-N118P-K121A-S122M | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[4-(2-azanylethylsulfamoyl)phenyl]pentanamide, ACETATE ION, IRIDIUM ION, ... | | Authors: | Hestericova, M, Heinisch, T, Alonso-Cotchico, L, Marechal, J.-D, Vidossich, P, Ward, T.R. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Directed Evolution of an Artificial Imine Reductase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6EUV
 
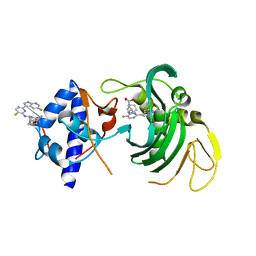 | |
6JGT
 
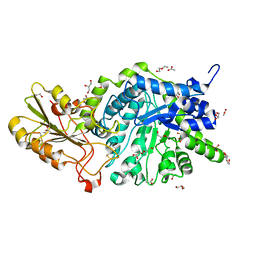 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
1JYK
 
 | |
1JYU
 
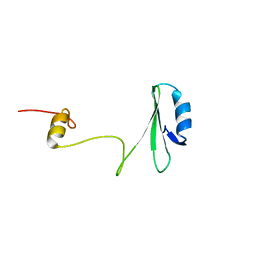 | | Xray Structure of Grb2 SH2 Domain | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
