5C9L
 
 | | Crystal structure of native PLL lectin from Photorhabdus luminescens at 1.65 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-27 | | Release date: | 2016-10-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
7CK6
 
 | | Protein translocase of mitochondria | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Yang, M, Wang, W, Zhang, L, Chen, X. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of human TOM core complex.
Cell Discov, 6, 2020
|
|
7LM2
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 3C | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Projected Dose Optimization of Amino- and Hydroxypyrrolidine Purine PI3K delta Immunomodulators.
J.Med.Chem., 64, 2021
|
|
7CI5
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (3R)-3-azanyl-4-oxidanylidene-4-[[3-(trifluoromethyloxy)phenyl]amino]butanoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CN0
 
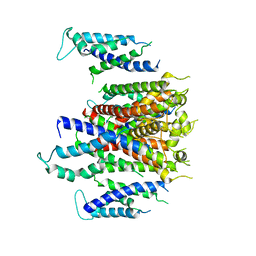 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
5BZ3
 
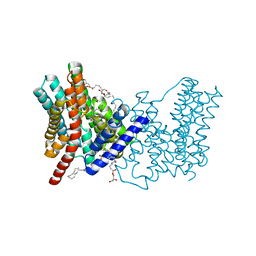 | | CRYSTAL STRUCTURE OF SODIUM PROTON ANTIPORTER NAPA IN OUTWARD-FACING CONFORMATION. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, Na(+)/H(+) antiporter | | Authors: | Coincon, M, Uzdavinys, P, Emmanuel, N, Cameron, A, Drew, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7C6K
 
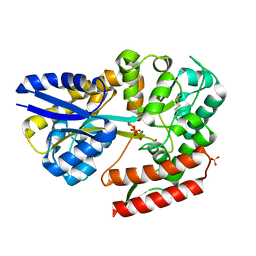 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotriose (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PYROPHOSPHATE 2-, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6W
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5C3E
 
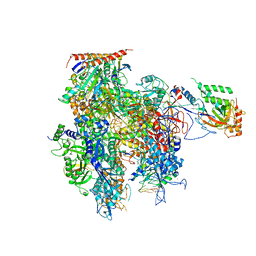 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5BZ6
 
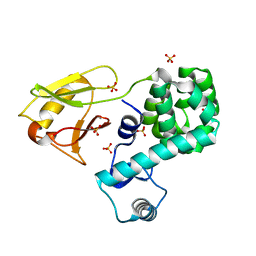 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
2ZSE
 
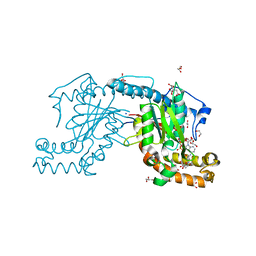 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP and Pantothenate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7CHK
 
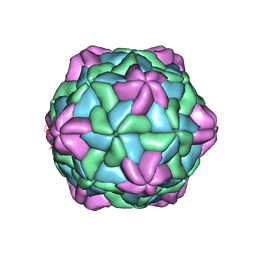 | | Cryo-EM Structure of Apple Latent Spherical Virus (ALSV) | | Descriptor: | VP20 protein, VP24 protein, VP25 protein | | Authors: | Naitow, H, Hamaguchi, T, Maki-Yonekura, S, Isogai, M, Yoshikawa, N, Yonekura, K. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Apple latent spherical virus structure with stable capsid frame supports quasi-stable protrusions expediting genome release.
Commun Biol, 3, 2020
|
|
5DZ9
 
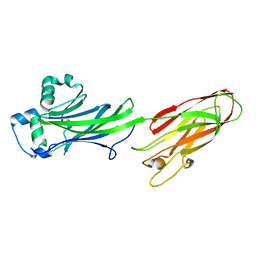 | |
5E2Q
 
 | | Structure of human DPP3 in complex with angiotensin-II | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kumar, P, Reisinger, M, Reithofer, V, Gruber, K. | | Deposit date: | 2015-10-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
7C66
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellobiose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7C6B
 
 | | Crystal structure of Ago2 MID domain in complex with 6-(3-(2-carboxyethyl)phenyl)purine riboside monophosphate | | Descriptor: | 3-[3-[9-[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]purin-6-yl]phenyl]propanoic acid, PHOSPHATE ION, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
1TF1
 
 | | Crystal Structure of the E. coli Glyoxylate Regulatory Protein Ligand Binding Domain | | Descriptor: | Negative regulator of allantoin and glyoxylate utilization operons | | Authors: | Walker, J.R, Skarina, T, Kudrytska, M, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-26 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical study of effector molecule recognition by the E.coli glyoxylate and allantoin utilization regulatory protein AllR.
J.Mol.Biol., 358, 2006
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C6L
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotriose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5E43
 
 | | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum | | Descriptor: | ACETATE ION, Beta-lactamase, NITRATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-05 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7095 Å) | | Cite: | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum
To Be Published
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
5E7K
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and cognate tRNALys in the A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
7CCS
 
 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
