4YKG
 
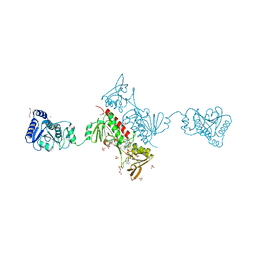 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NAD+ from Escherichia coli | | Descriptor: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
5MNW
 
 | |
7EI2
 
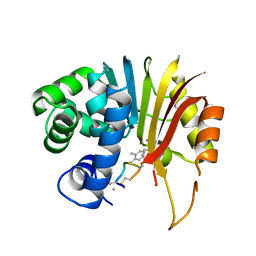 | | Structure of human NNMT in complex with macrocyclic peptide 8 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 8 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
5CJ3
 
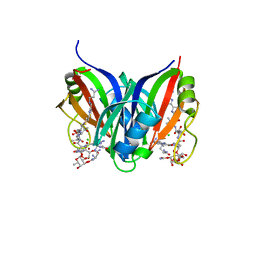 | | Crystal structure of the zorbamycin binding protein (ZbmA) from Streptomyces flavoviridis with zorbamycin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Zbm binding protein, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Rudolf, J.D, Ma, M, Chang, C.-Y, Lohman, J.R, Yang, D, Shen, B, Enzyme Discovery for Natural Product Biosynthesis, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-13 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
7EFH
 
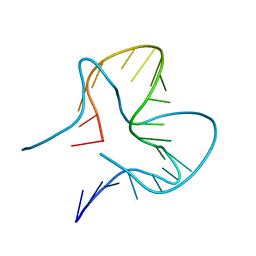 | | RNA kink-turn motif | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP*GP*AP*GP*CP*C)-3') | | Authors: | Kondo, J, Nagashima, M. | | Deposit date: | 2021-03-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RNA kink-turn motif
To Be Published
|
|
331D
 
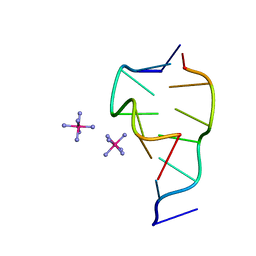 | | CRYSTAL STRUCTURE OF D(GCGCGCG) WITH 5'-OVERHANG G'S | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*GP*CP*GP*CP*G)-3') | | Authors: | Pan, B, Ban, C, Wahl, M, Sundaralingam, M. | | Deposit date: | 1997-05-13 | | Release date: | 1997-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of d(GCGCGCG) with 5'-overhang G residues.
Biophys.J., 73, 1997
|
|
5MXL
 
 | |
5MWW
 
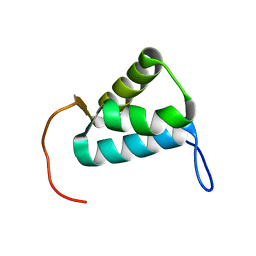 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
5COE
 
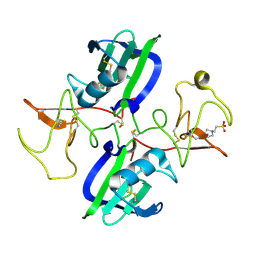 | | The structure of the NK1 fragment of HGF/SF complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Ascher, D.B, Chirgadze, D.Y, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-20 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5MZF
 
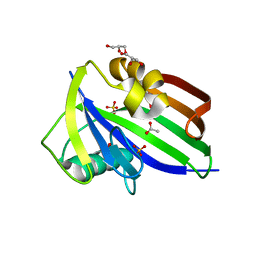 | | Crystal structure of dog MTH1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5MZQ
 
 | | X-ray structure of the M205W mutant of GLIC in complex with bromoform | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-28 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
5BYY
 
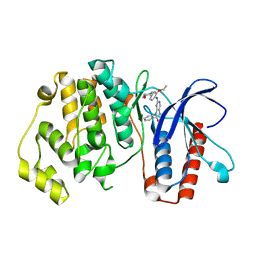 | | ERK5 IN COMPLEX WITH SMALL MOLECULE | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5BZ2
 
 | |
5N0M
 
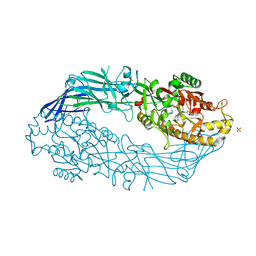 | | hPAD4 crystal complex with BB-F-amidine | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, SULFATE ION, ... | | Authors: | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | Deposit date: | 2017-02-03 | | Release date: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
5C92
 
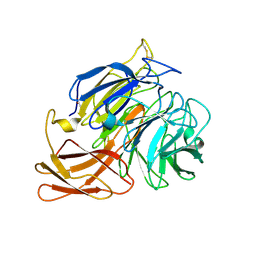 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, in complex with copper | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (I) ION, ... | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
3VC0
 
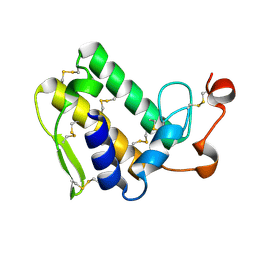 | | Crystal structure of Taipoxin beta subunit isoform 1 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino de Laureto, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
5CBV
 
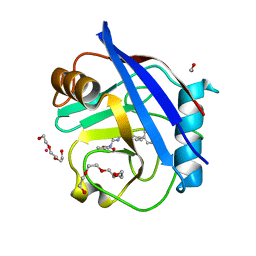 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, Human Cyclophilin D, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
7ELK
 
 | |
5C3C
 
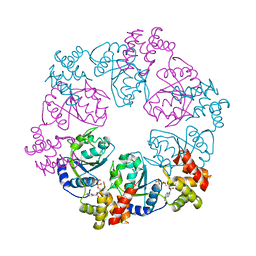 | |
7EFG
 
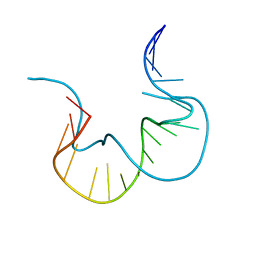 | | RNA kink-turn motif | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP*GP*AP*GP*CP*C)-3') | | Authors: | Kondo, J, Nagashima, M. | | Deposit date: | 2021-03-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA kink-turn motif
To Be Published
|
|
5C4P
 
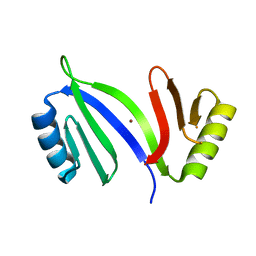 | |
7EIF
 
 | | Crystal structure of GAS41 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 4 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7EKU
 
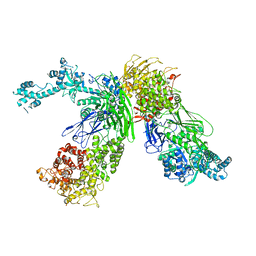 | |
7EG5
 
 | |
5CG5
 
 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.402 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
