8ONT
 
 | | Structure of Setaria italica NRAT in complex with a nanobody | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, NRAMP related aluminium transporter, Nanobody1 | | Authors: | Ramanadane, K, Liziczai, M, Markovic, D, Straub, M.S, Rosalen, G.T, Udovcic, A, Dutzler, R, Manatschal, C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural and functional properties of a plant NRAMP-related aluminum transporter.
Elife, 12, 2023
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
4CVW
 
 | | Structure of the barley limit dextrinase-limit dextrinase inhibitor complex | | Descriptor: | CALCIUM ION, LIMIT DEXTRINASE, LIMIT DEXTRINASE INHIBITOR | | Authors: | Moeller, M.S, Vester-Christensen, M.B, Jensen, J.M, Abou Hachem, M, Henriksen, A, Svensson, B. | | Deposit date: | 2014-03-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal Structure of Barley Limit Dextrinase:Limit Dextrinase Inhibitor (Ld:Ldi) Complex Reveals Insights Into Mechanism and Diversity of Cereal-Type Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4BWO
 
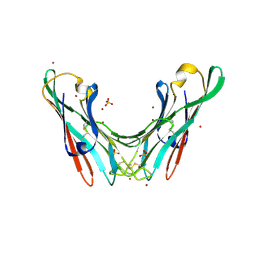 | | The FedF adhesin from entrrotoxigenic Escherichia coli is a sulfate- binding lectin | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Lonardi, E, Moonens, K, Buts, L, de Boer, A.R, Olsson, J.D.M, Weiss, M.S, Fabre, E, Guerardel, Y, Deelder, A.M, Oscarson, S, Wuhrer, M, Bouckaert, J. | | Deposit date: | 2013-07-03 | | Release date: | 2013-08-28 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia Coli
Biology, 2, 2013
|
|
8ANP
 
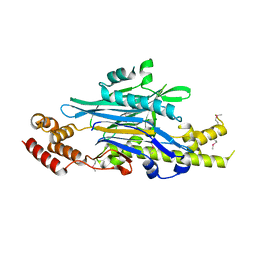 | | Legionella effector Lem3 mutant D190A in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, Phosphocholine hydrolase Lem3, SULFATE ION, ... | | Authors: | Kaspers, M.S, Pogenberg, V, Ernst, S, Ecker, F, Pett, C, Ochtrop, P, Hedberg, C, Groll, M, Itzen, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
6I5P
 
 | |
6I68
 
 | |
1ID8
 
 | | NMR STRUCTURE OF GLUTAMATE MUTASE (B12-BINDING SUBUNIT) COMPLEXED WITH THE VITAMIN B12 NUCLEOTIDE | | Descriptor: | 2-HYDROXY-PROPYL-AMMONIUM, METHYLASPARTATE MUTASE S CHAIN, PHOSPHORIC ACID MONO-[5-(5,6-DIMETHYL-BENZOIMIDAZOL-1-YL)-4-HYDROXY-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER | | Authors: | Tollinger, M, Eichmuller, C, Konrat, R, Huhta, M.S, Marsh, E.N.G, Krautler, B. | | Deposit date: | 2001-04-04 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum traps the nucleotide moiety of coenzyme B(12).
J.Mol.Biol., 309, 2001
|
|
7MLK
 
 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
6BDU
 
 | |
8B8K
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8G
 
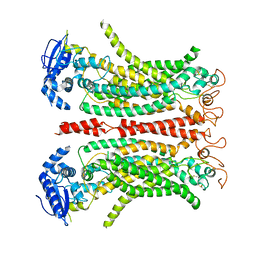 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6 | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8J
 
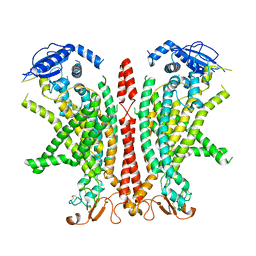 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC1
 
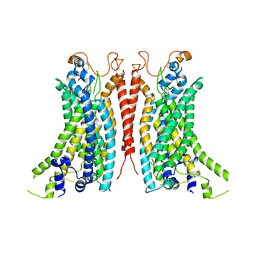 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
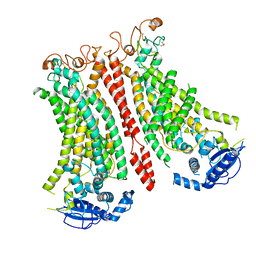 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
2BHL
 
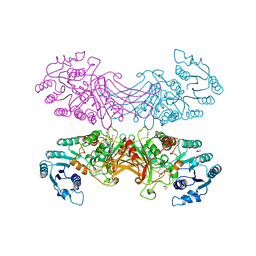 | | X-RAY STRUCTURE OF HUMAN GLUCOSE-6-PHOSPHATE DEHYDROGENASE (DELETION VARIANT) COMPLEXED WITH GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL | | Authors: | Kotaka, M, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Studies of Glucose-6-Phosphate and Nadp+ Binding to Human Glucose-6-Phosphate Dehydrogenase
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8B8Q
 
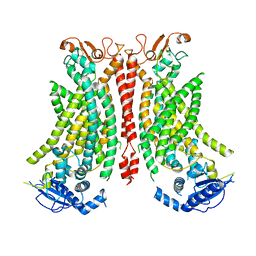 | | Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
6CIL
 
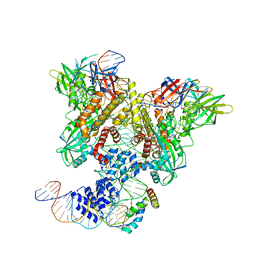 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIK
 
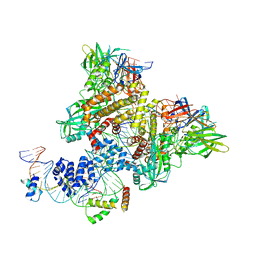 | | Pre-Reaction Complex, RAG1(E962Q)/2-intact/nicked 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 12RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
5UNH
 
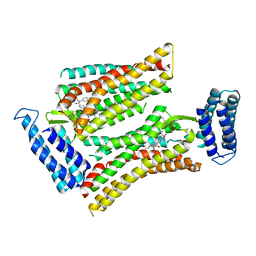 | | Synchrotron structure of human angiotensin II type 2 receptor in complex with compound 2 (N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'- biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide) | | Descriptor: | N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide, Soluble cytochrome b562,Type-2 angiotensin II receptor | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
6CIM
 
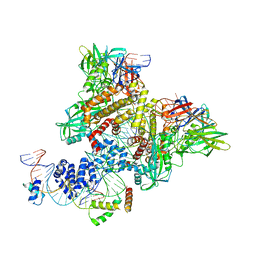 | | Pre-Reaction Complex, RAG1(E962Q)/2-nicked/intact 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 23RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6I7A
 
 | |
7O0T
 
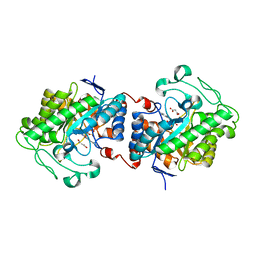 | | Crystal structure of Chloroflexus aggregans ene-reductase CaOYE holoenzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, MALONATE ION, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Robescu, M.S, Niero, M, Loprete, G, Bergantino, E, Cendron, L. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A New Thermophilic Ene-Reductase from the Filamentous Anoxygenic Phototrophic Bacterium Chloroflexus aggregans .
Microorganisms, 9, 2021
|
|
