7YDS
 
 | | The structure of the bispecific antibody targeted PD-L1 and 4-1BB | | Descriptor: | Anti-PDL1-VH-CH1, Anti-PDL1-VL-CL, Programmed cell death 1 ligand 1 | | Authors: | Gao, Y, Zhu, M, Liu, W.T, Cheng, L.S, Zhu, Z.L, Niu, L.W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bispecific antibody targeted PD-L1 and 4-1BB induces a potent antitumor immune activity in colorectal cancer without systemic toxicity
To Be Published
|
|
7YHN
 
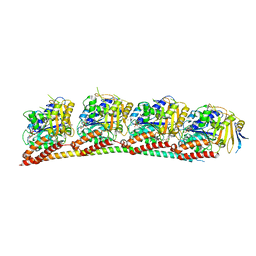 | | ANTI-TUMOR AGENT Y48 IN COMPLEX WITH TUBULIN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methyl-3-[(4-methylphenyl)sulfonylamino]-~{N}-[(6-methylpyridin-3-yl)methyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Du, T, Ji, M, Hou, Z, Lin, S, Zhang, J, Wu, D, Zhang, K, Lu, D, Xu, H, Chen, X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site.
J.Med.Chem., 65, 2022
|
|
3B76
 
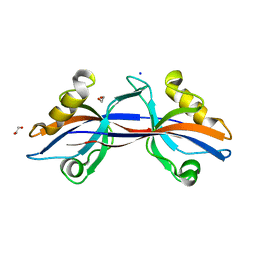 | | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase LNX, SODIUM ION | | Authors: | Ugochukwu, E, Burgess-Brown, N, Berridge, G, Elkins, J, Bunkoczi, G, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Gileadi, O, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
To be Published
|
|
3B6P
 
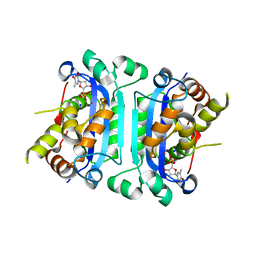 | | Structure of TREX1 in complex with a nucleotide and inhibitor ions (sodium and zinc) | | Descriptor: | SODIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1, ... | | Authors: | Brucet, M, Querol-Audi, J, Fita, I, Celada, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical studies of TREX1 inhibition by metals. Identification of a new active histidine conserved in DEDDh exonucleases.
Protein Sci., 17, 2008
|
|
2B37
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-octyl-2-phenoxyphenol | | Descriptor: | 5-OCTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
3BA7
 
 | |
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | Descriptor: | D-Pep-1, L-19437 | | Authors: | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
3BC1
 
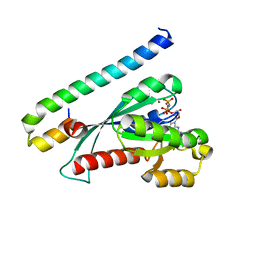 | | Crystal Structure of the complex Rab27a-Slp2a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A, ... | | Authors: | Chavas, L.M.G, Ihara, K, Kawasaki, M, Wakatsuki, S. | | Deposit date: | 2007-11-12 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a
Structure, 16, 2008
|
|
3BDU
 
 | | Crystal structure of protein Q6D8G1 at the resolution 1.9 A. Northeast Structural Genomics Consortium target EwR22A. | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein Q6D8G1 at the resolution 1.9 A.
To be Published
|
|
7EDS
 
 | |
7EEU
 
 | |
3BFA
 
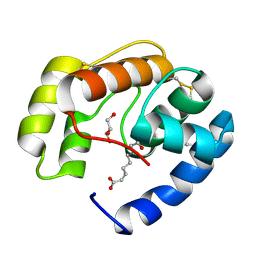 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3BBR
 
 | |
6JV6
 
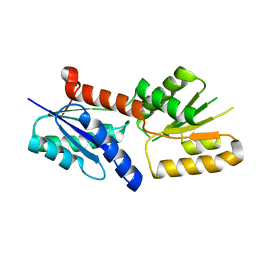 | | Crystal structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subspecies spizizenii in complex with cobalt | | Descriptor: | COBALT (II) ION, Sirohydrochlorin ferrochelatase | | Authors: | Nam, M.S, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2019-04-16 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cobalt complex structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subsp. spizizenii.
KOREAN J MICROBIOL., 55, 2019
|
|
7DTI
 
 | |
7VYN
 
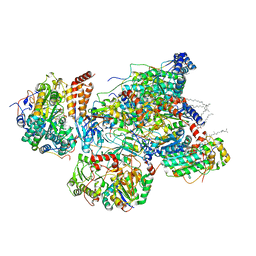 | | Matrix arm of active state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-14 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VZ8
 
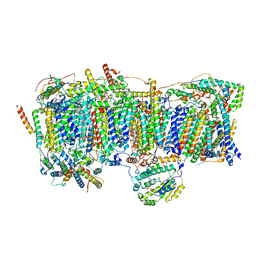 | | Membrane arm of deactive state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VZ1
 
 | | Matrix arm of deactive state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7E5P
 
 | | Aptamer enhancing peroxidase activity of myoglobin | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*AP*GP*GP*G)-3') | | Authors: | Tsukakoshi, K, Matsugami, A, Khunathai, K, Kanazashi, M, Yamagishi, Y, Nakama, K, Oshikawa, D, Hayashi, F, Kuno, H, Ikebukuro, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol.
Nucleic Acids Res., 49, 2021
|
|
7EN4
 
 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
7EAU
 
 | | SIB1, an effector of Colletotrichum orbiculare | | Descriptor: | SIN1 | | Authors: | Mori, M, Ohki, S, Kurita, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-11-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fungal effector SIB1 of Colletotrichum orbiculare has unique structural features and can suppress plant immunity in Nicotiana benthamiana.
J.Biol.Chem., 297, 2021
|
|
3AZV
 
 | | Crystal structure of the receptor binding domain | | Descriptor: | D/C mosaic neurotoxin, SULFATE ION | | Authors: | Nuemket, N, Tanaka, Y, Tsukamoto, K, Tsuji, T, Nakamura, K, Kozaki, S, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism
Biochem.Biophys.Res.Commun., 411, 2011
|
|
6I4S
 
 | | Crystal structure of the disease-causing R447G mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Hubert, A, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
3B0H
 
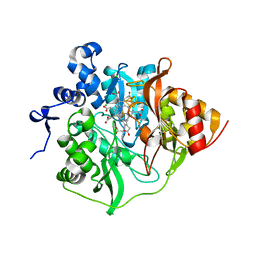 | | Assimilatory nitrite reductase (Nii4) from tobbaco root | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
7EH3
 
 | |
